Erik Vidholm, Ewert Bengtsson, Ingela Nyström, Stefan Seipel
Funding: Swedish Research Council
Period: 0301-
Partners: Lennart Thurfjell, GE Healthcare, Uppsala/London, UK; Gunnar Jansson, Dept. of Psychology, UU; Hans Frimmel, Dept. of Oncology, Radiology, and Clinical Immunology, UU
Abstract: Modern medical imaging techniques provide 3D images of increasing complexity. Better ways of exploring these images for diagnostic and treatment planning purposes are needed. Combined stereoscopic and haptic display of the images form a powerful platform for such image analysis.
In order to work with specific patient cases it is necessary to be able to work directly with the medical image volume and to generate the relevant 3D structures directly as they are needed for the visualization. Most work so far on haptic display has used predefined object surface models. In this project we are creating the tools necessary for effective interactive exploration of complex medical image volumes for diagnostic or treatment planning purposes through combined use of haptic and 3D stereoscopic display techniques. The project is based on an evaluation of previous approaches from literature and our own previous experiences. The developed methods are tested on real medical application data.
During 2004, we have developed a fast surface renderer for interactive modeling and manipulation of image data with haptic feedback. We have also developed a haptic interaction method based on gradient diffusion for enhanced navigation. See Projects 14 and 15.
Erik Vidholm, Jonas Agmund, Ewert Bengtsson
Funding: Swedish Research Council
Period: 0401-
Abstract: In this project, we have developed a haptic-enabled application for interactive editing in medical image segmentation. We use a fast surface rendering algorithm based on marching cubes (MC) to display different segmented objects, and we apply a proxy-based volume haptics algorithm to be able to touch and edit these objects at interactive rates. As an application example, we show how the system can be used to initialize a fast marching segmentation algorithm for extracting the liver in magnetic resonance (MR) images and then edit the result if it is incorrect, see Figure 5.
![\includegraphics[width=5.2cm]{Images/screenshot1.ps}](img27.png) ![\includegraphics[width=5.2cm]{Images/screenshot2.ps}](img28.png) ![\includegraphics[width=5.2cm]{Images/screenshot3.ps}](img29.png) |
The project results are documented in the MSc thesis by Agmund, see Section 3.3, and in a conference publication that was presented at SIGRAD'04, Gävle, Sweden.
Erik Vidholm, Xavier Tizon, Ingela Nyström, Ewert Bengtsson
Period: 0301-
Funding: Swedish Research Council; SLU S-faculty; (This project was previously part of the Swedish Foundation for Strategic Research VISIT programme)
Abstract: The manual step in semi-automatic segmentation of medical volume images typically involves initialization procedures such as placement of seed-points or positioning of surface models inside the object to be segmented. The initialization is then used as input to an automatic algorithm. We investigate how such initialization tasks can be facilitated by using haptic feedback.
By using volume haptics we aim to make the initialization process easier and more efficient. In April 2004, we presented a haptic-enhanced seeding method for magnetic resonance angiography (MRA) images in order to separate arteries and veins at the IEEE International Symposium on Biomedical Imaging (ISBI'04). The haptic interaction was based on the volume gradient and made it possible to trace vessels by sliding against the inner vessel wall.
During 2004, we also started a project for interactive segmentation of MR images of the liver as a step in surgery planning. To be able to work with larger objects such as the liver we base the haptic interaction on gradient vector flow (GVF) that propagates edge information from strong boundaries into the center of objects. This approach allows a user to feel object boundaries while still being centered inside the object. The method is not limited to elongated structures such as vessels. Initial results are documented in a paper that will be presented at the conference WorldHaptics in March 2005, Pisa, Italy.
Felix Wehrmann, Ewert Bengtsson
Funding: UU TN-faculty
Period: 9912-0405
Abstract: This project started under the scope of the general idea of model-based segmentation. A large number of images, especially from the medical sector, lack a proper description of the objects the image analyst is interested in. Often, this leads to poor results of automated segmentation procedures, if any. Incorporating information about the character of an object, like shape information or colour appearance, is one possible completion of a description. However, many models that are used for description lack the property to compensate for the variation nature supplies us with. As an example, we could ask ourselves, which features make us easily recognise and localise a brain in a medical 3D-image, a task which has automated solutions only in specific cases.
With the intention to compensate for natural variation, we applied a number of common concepts to the problem. In particular, orthogonal transforms, such as PCA and ICA, have been inspected in an attempt to derive the characteristic correlations between similar shapes. Moreover, the applicability of Markov random fields as a local stochastic modeling concept was analysed.
It turned out that a general model should not be dependent on landmarks as required for the previous transformations. It has been realised that the avoidance of landmarks place shape data on a ground comparable to images. Since variations in landmark-less shape data appear as non-linear manifolds, as well as the appearance change of colour in images, a neural network was designed to acquire the particularities of the data. After training on examples, the network provides a non-linear representation of object appearance by means of its modes of variation. Neural networks have been found a promising representation for the explored kind of variation in the data. They were applied to a variety of data and their learning abilities have been studied further. This project lead to a PhD for Wehrmann, see Section 4.2.
Hamed Hamid Muhammed
Funding: UU TN-faculty, Swedish National Space Board
Period: 0201-
Abstract: New neuro-fuzzy systems (Weighted Neural Networks, WNN) which can characterize the distribution of a given data set were developed in this work. The basic idea is based on the famous Hebb's postulate which states that the connection between two winning neurons gets stronger. The WNN algorithm produces a net of nodes connected by edges. Additional weights, which are proportional to the local densities in input space, are associated with the resulting nodes and edges to store useful information about the topological relations in the given input data set. A fuzziness factor, proportional to the connectedness of the net, is introduced in the system. The resulting net reflects and preserves the topology of the input data set, and can be considered as a fuzzy representation of the data set. Two main types of WNNs were developed: incremental self-organising and fixed (grid-partitioned) depending on the underlying ANN algorithm.
- Weighted Fixed Neural Networks (WFNN): A number of zero-weighted nodes are uniformly distributed in input space where the given data set is found. Then, weights are assigned to these nodes, where a relatively higher node-weight corresponds to a relatively denser region of the data set. Weighted connections are established between neighbouring nodes, where the weights are also proportional to the local density of input data. The work has resulted in a journal paper presenting the WFNN algorithm.
- Weighted Incremental Neural Networks (WINN): The model is built by
successive addition, adaptation, and sometimes deletion of elements
(i.e., nodes and edges), according to suitable strategies, until a
stopping criterion is met. Here also, a weighted connected net, which
preserves the topology of the input data set, is produced. The algorithm
begins with only two nodes connected by an edge, then new nodes and
edges are generated and the old ones are updated (and
sometimes deleted) while the learning process proceeds until a certain
stopping criterion is met. The work has resulted in a journal paper (2004) presenting the WINN algorithm and a reviewed conference paper (2003) comparing WINN with WFNN.
Ola Weistrand, Gunilla Borgefors
Funding: Swedish Research Council; UU TN-faculty
Period: 9701-
Partners: Christer Kiselman, Dept. of Mathematics, UU; Örjan Smedby, Dept. of Medicine and Care, Linköping University Hospital
Abstract: Shape description derived from volume images is usually local, e.g., finite elements, surface facets, and spline functions. This can be a severe limitation on usefulness, as comparison between different shapes becomes very difficult. In 2D, Fourier descriptors is a successful and often used global descriptor with adaptable accuracy. This concept cannot be immediately generalised to 3D because it relies heavily on the existence of an ordering of the boundary pixels. The aim of this project is to overcome this problem and develop methods for global shape description in 3D. At the moment we study a limited class of objects, those that are homotopic to the sphere. By using harmonic functions we map the object's surface onto the sphere and correct distortions resulting from this mapping by non-linear optimization methods. Shape invariants can then be calculated using spherical harmonic functions.
Gunilla Borgefors, Ida-Maria Sintorn, Stina Svensson
Funding: SLU S-faculty
Period: 9309-
Abstract: In a distance transform (DT), each picture element in an object is labeled with the distance to the closest element in the background. Thus the shape of the object is ``structured" in a useful way. Only local operations are used, even if the results are global distances. DTs are very useful tools in many types of image analysis, from simple noise removal to advanced shape recognition. We have investigated DTs since the early 1980's.
In medical and industrial volume images, the picture elements are often rectangular boxes rather than cubes, because the images are created as a stack of ``slices'', either physically or by computation. It would be time and memory saving to work directly in such grids, rather than first interpolating the image to a cubic grid. However, DT based methods (among others) are not directly applicable to non-cubic grids. Therefore, we have investigated DTs in box grids, where voxels have two equal sides and the third is longer. A voxel in such a grid has five different types of neighbours, for which optimal weights were calculated as a function of the voxel elongation. The optimisation in 3D gives rise to four types of regular DTs of which one, the simplest, was further investigated. An article with the results was published in Pattern Recognition Letters vol 25, 2004.
Robin Strand, Gunilla Borgefors
Funding: Graduate School in Mathematics and Computing (FMB)
Period: 0308-
Partners: Christer Kiselman, Dept. of Mathematics, UU
Abstract: Volume images are usually captured in one of two ways: either the object is sliced (mechanically or optically) and the slices put together into a volume or the image is computed from raw data, such as X-ray or magnetic tomography. In both cases, voxels are usually box-shaped, as the within slice resolution is higher than the between slice distance. Before applying image analysis algorithms, the images are usually interpolated into the cubic grid. However, the cubic grid might not be the best choice. In two dimensions, it has been demonstrated in many ways that the hexagonal grid is theoretically better than the square grid. The body-centered cubic (bcc) grid and the face-centered cubic (fcc) grid are the generalizations to 3D of the hexagonal grid. In the bcc grid the voxels consist of truncated octahedra, in the fcc grid the voxels consist of rhombic dodecahedra. The voxels in these grids are better approximations to Euclidean balls than the cube, a fact that is justified by looking at the voxels neighbours. A voxel in the fcc grid has as many as twelve first neighbours and thus constitutes the densest periodic packing of the grids. A voxel in the bcc grid has 14 face neighbours, of which eight are first neighbours.
The main goal of the project is to develop image analysis and processing methods for volume images digitized in the fcc and bcc grids, especially distance transform based and morphological methods for shape description and analysis. Weighted distance transforms, see also Project 19, in the fcc and bcc grids have been investigated and the results have been submitted. A skeletonization algorithm based on iterative, topology preserving, thinning of distance transformed objects in the fcc and bcc grids has been developed. The results were presented at the ICPR conference.
Nataša Sladoje (Matic), Ingela Nyström, Joakim Lindblad, Gunilla Borgefors
Funding: SLU S-faculty, UU TN-faculty
Period: 0109-
Partners: Punam K. Saha, MIPG, Dept. of Radiology, University of Pennsylvania, Philadelphia, USA; Jocelyn Chanussot, Signal and Image Laboratory (LIS), INPG, Grenoble, France
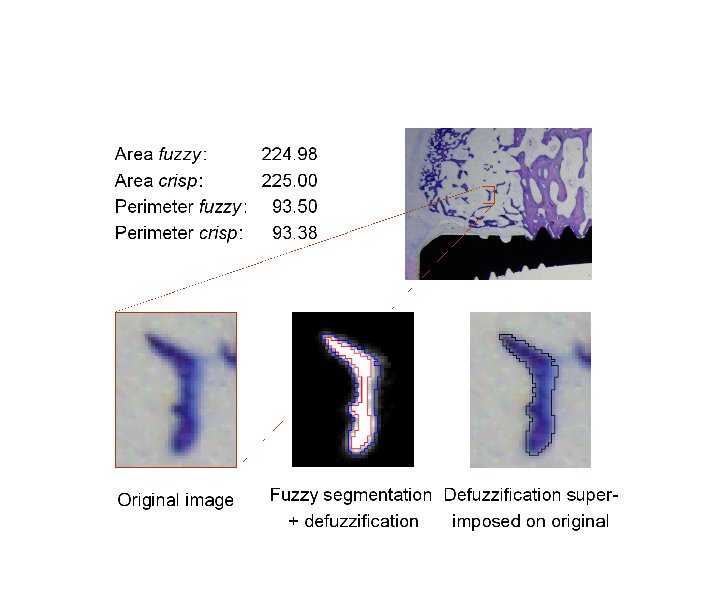
Abstract: Fuzzy segmentation methods, that have been developed in order to reduce the negative effects of the unavoidable loss of data in the digitisation process, initialise the interest for new shape analysis methods, handling grey-level images. We assume that in the segmentation process most picture elements easily can be classified either as object or background, but for elements in the vicinity of the boundary of the digitised object it is hard to make such a discrimination. One way to treat such an element is to determine the extent of its membership to the object as the fraction of its area that belongs to the original object. We have performed studies on perimeter and area of 2D, and surface area and volume of 3D fuzzy subsets, where the focus has been on objects with fuzzy borders. We have implemented a method where we propose perimeter and area estimators, as well as surface area and volume estimators, adjusted to the discrete case. We have concluded that our method highly improves both accuracy and precision of the results obtained from crisp (hard) segmentation, especially in the case of low resolution images, i.e., small objects.
During 2004, our defuzzification method based on feature invariance has been further developed. The idea is to use, not only membership and/or gradient information, which is a common approach, but also shape information, in order to generate a crisp representation of a fuzzy shape. Having precise area and perimeter estimates, we incorporated them, together with the centre of gravity of a shape, into a defuzzification method. We have developed and tested a number of optimization algorithms for finding the crisp object which most resembles the observed fuzzy object. Having a computationally demanding task, we try to provide a balance between speed and quality/precision of the procedures. Our method can be useful whenever it is important to preserve reliable measures of the original object, along the object analysis procedure. An example of a defuzzification of a region close to a bone implant, by using the suggested method, is shown in Figure 6.
Following the main goal of extending various shape descriptors to the fuzzy case, we analysed different ways of computing the signature of a fuzzy shape, based on the distance from the shape centroid. The encouraging results advocate use of fuzzy shapes to improve precision of the descriptor, especially at low resolutions. These results will be published in the journal Pattern Recognition Letters in early 2005.
Ingela Nyström, Gunilla Borgefors
Funding: UU TN-faculty, SLU S-faculty
Period: 0209-
Partners: Gabriella Sanniti di Baja, Istituto di Cibernetica, CNR, Pozzuoli, Italy
Abstract: 2D grey-level images are interpreted as 3D binary images, where the grey-level plays the role of the third coordinate. In this way, algorithms devised for 3D binary images can be used to analyse 2D grey-level images without prior segmentation into hard object and background. One algorithm computes an approximation of the convex hull of the 2D grey-level object, by building a covering polyhedron closely fitting the corresponding object in a 3D binary image. The obtained result is convex both from a geometrical and a grey-level point of view. Another algorithm skeletonizes a 2D grey-level object by skeletonizing the top surface of the object in the corresponding 3D binary image. The obtained 3D curve skeleton is pruned, before being projected back to a 2D grey-level image. This is suitably post-processed, since the projection may cause spurious loops and thickening. This algorithm can find applications in optical character recognition (OCR) and document analysis or in other situations where shape analysis by skeletons is desired. For an example, see Figure 7.
![\includegraphics[width=0.25\textwidth]{/usr/users/ingela/public_html/Grey2DSkeleton/eight.ps}](img31.png)
![\includegraphics[width=0.25\textwidth]{/usr/users/ingela/public_html/Grey2DSkeleton/UppsalaDec02/eightunzig3D.ps}](img32.png)
![\includegraphics[width=0.25\textwidth]{/usr/users/ingela/public_html/Grey2DSkeleton/UppsalaDec02/eightunzig.ps}](img33.png)
|
Stina Svensson
Funding: SLU S-faculty; CNR Italy
Period: 9801-
Partners: Carlo Arcelli and Gabriella Sanniti di Baja, Istituto di Cibernetica, CNR, Pozzuoli, Italy
Abstract: When analysing the shape of an object it is not only of interest to study the object itself but also its complement. This can give us information on, for example, the structure of tunnels possibly existing in the object. We start the analysis by identifying the convex deficiency of the object, i.e., the difference between the convex hull of the object and the object. The convex deficiency can then be decomposed into regions corresponding to cavities or tunnels in the object. The structure of the tunnels can be further analysed with respect to branching, thickness, and length. An article describing this analysis in detail is published in Image and Vision Computing in February 2005.
Stina Svensson
Funding: SLU S-faculty
Period: 0210-
Partners: Pieter Jonker, Pattern Recognition Section, Faculty of Applied Physics, Delft University of Technology, Delft, The Netherlands
Abstract: In medical applications, devices giving three dimensional (3D) images are in use since the sixties for imaging anatomical or functional aspects of the human body. It is also possible to use images with more than three dimensions. Four dimensional (4D) images can be used to represent a 3D object traversing in space-time, e.g., a beating heart or a moving robot. We have started to analyse the shape of the objects in such images. As 4D images contain huge amounts of data, we first need to find a suitable representation with a reduced amount of data, still containing enough information to actually perform shape analysis. Skeletons is such a representation (see Projects 22,4), were dimension is reduced one or several steps. We have started the work on developing topologically correct 4D skeletonization methods.