Maria Axelsson, Ida-Maria Sintorn, Stina Svensson, Gunilla Borgefors
Funding: SLU S-faculty
Period: 0406-
Partners: STFI-Packforsk, Stockholm, StoraEnso, Falun, Norwegian Pulp and Paper Research Institute (PFI), Trondheim, Norway
Abstract: The internal structure of paper is important to study since many paper properties correspond directly to the properties of single fibres and their interaction in the fibre network. How single fibres in paper bond and how this effects paper quality is not fully understood, since most structure analysis of paper has been done in cross sectional 2D images and paper is a complex 3D structure. Image analysis of 3D images of paper and development of measurements of network properties and individual fibre properties can be a great contribution to the development in this area.
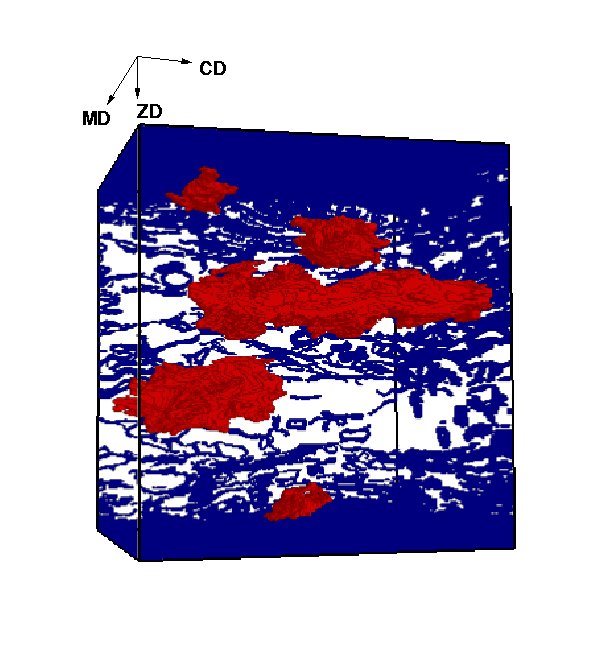
In this project, both segmentation algorithms for individual fibres and for the porous structure are investigated, see Figure 2 for individually segmented pores. The project objective is to achieve a fully segmented paper volume where any measurement of the internal structure is available, creating relations between microscopical and macroscopical properties of paper. These methods can also be used for other fibrous and porous materials.
In the project a volume image of paper, created from a series of 2D scanning electron microscopy (SEM) images at StoraEnso in Falun, is available for the studies. Other volume images are also used and new methods for creating other sample volume images are investigated.
Mats Erikson, Gunilla Borgefors
Funding: SLU S-faculty
Period: 9508-0411
Partners: Tomas Brandtberg; Kenneth Olofsson, Dept. of Forest Resource Management and Geomatics, SLU, Umeå
Abstract: The main goal of the project is to develop methods for computerised analysis of high spatial resolution remotely sensed data, i.e., digitised aerial photographs and laser scanning data, and to use the results in forestry and environmental assessment instead of (or as a complement to) field visits by humans. A set of 50 research aerial images (digitised colour-IR film), with resolution approximately 10 cm and 3 cm (flight height 600 m, focal length 300 mm) to make the individual tree crowns clearly visible is used. Interesting forest stand parameters to measure in the images are: number and positions of trees, horizontal tree crown areas, tree heights, and tree species composition. Features related to the individual tree species are, e.g., colour, internal structure (texture), and boundary structure. Algorithms for very precise segmentation of individual tree crowns have been developed during the years. Also an algorithm for classification of the tree crowns into the tree species, birch, aspen, spruce, and pine has been developed. On the stand level, the classification results are 95% correct. One of the segmentation methods has also been used to segment tree crowns in tropical forest with promising results. A new thesis has come out from the project, see Section 4.2.
Hamed Hamid Muhammed
Funding: UU TN-faculty, Swedish National Space Board
Period: 0201-
Partners: Anders Larsolle, Dept. of Biometry and Engineering, SLU, Uppsala
Abstract: The impact of plant pathological stress on crop reflectance can be measured both in broad band vegetation indices and in narrow or local characteristics of the reflectance spectra. Our goal is to use the whole spectra in the objective examination of how different parts of the spectrum contribute in describing disease severity in wheat. A reference data set is first collected, consisting of hyper spectral crop reflectance data vectors and the corresponding disease severity field assessments. Two approaches were addressed to achieve this goal:
- A hyper spectral reflectance spectrum was considered as a mixed signal, i.e. the integration of the effects of all active objects in the investigated area. Independent component analysis (ICA) was used to blindly separate mixed statistically independent signals. Principal component analysis (PCA) was also used to extract interesting components. The ICA or PCA results had then to be interpreted efficiently. This was achieved by using a technique called Feature Vector Based Analysis (FVBA), which produces a number of ``component-feature vector'' pairs, which represent the spectral signatures and the corresponding weight coefficients of the different constituting source signals. High correlations were found between these weight coefficients and the corresponding field assessments of disease severity in the crop. The effects of increased disease severity could be easily observed from the resulting disease-specific signatures.
- The hyper spectral vectors, from the reference data set, are first normalised into zero-mean and unit-variance vectors by performing various combinations of spectral- and band-wise normalisations. Then, after applying the same normalisation procedures to the new hyper spectral data that we wish to analyse, a nearest neighbour classifier is used to classify the new data against the reference data. Finally, the corresponding signatures, that describe the studied stress, are computed using a linear transformation model. High correlation is obtained between the classification results and the corresponding disease severity, confirming the usefulness and efficiency of this approach. The low computational load of this approach makes it suitable for real-time on-vehicle applications. A paper about this topic has been submitted for publication.
Ida-Maria Sintorn, Magnus Gedda, Stina Svensson, Gunilla Borgefors
Funding: SLU S-faculty; UU TN-faculty
Period: 0401-
Partners: Susana Mata, Rey Juan Carlos University, Madrid, Spain; Sidec Technologies AB, Stockholm
Abstract: Sidec Electron Tomogprahy (SET) is a 3D protein imaging method which produces volumes with a resolution of a few . This is good enough to give coarse structural information about the shape of proteins and how they interact with other proteins. The shape information available is, however, limited as the proteins only consist of a few thousand voxels, which is a small amount when it comes to 3D analysis. We develope several methods related to the analysis of 3D images of proteins, aqcuired using SET or reconstructed from atom positions available from the protein data bank.
The SET volumes contain hundreds of objects of which true proteins are visually identified after grey-level thresholding of the volume and size discrimination of connected components. The visual inspection is very time consuming and varying background makes it difficult to find a suitable threshold. Proteins can also, after thresholding, consist of several disconnected parts or be connected to other objects. The consequence of this is that true proteins are easily discarded by the size discimination step. During 2004 a method which combines a stable edge detection algorithm and contour based template matching was developed to identify objects in the volumes resembling the true protein. The method reduces the amount of visual inspection needed and adresses the problems of varying background and size discrimination. It shows very promising results but need to be evaluated on more SET volumes.
To facilitate the recognition and analysis of proteins in volume images, two representation schemes are being developed. One way of representing an object is to use decomposition into significant parts. Object recognition can then be seen as a hierarchical process. Each part can be analysed individually as well as how it is connected to other subparts to create the complete object. During 2003, a decomposition method, also developed at CBA and designed for binary images, was further developed to incorporate grey-level information and adapted to the application of protein decomposition. This method was presented at the International Symposium on Biomedical Imaging (ISBI) 2004.
Another way of representing an object is by a medial skeletal-like curve. A medial surface representation extracted directly from the grey-level image has earlier been developed at CBA. This representation differs from other surface skeletons by prioring grey-level to shape. A great advantage with this approach is that no segmentation of the object is needed prior to extracting the representation. Since the grey-level distribution within a protein is of great importance for function this medial representation was adapted to suit proteins in volume images during 2004 giving a curve connecting the locii of the different parts of the protein as a medial representation. The representation serves as a complement to the protein decomposition method. This protein medial grey-level based representation will be presented at the IbPRIA conference in June 2005. In Figure 3, the two representation schemes are shown on three antibody proteins imaged using SET.

|
Ida-Maria Sintorn, Gunilla Borgefors
Funding: SLU S-faculty
Period: 0111-
Partners: Mohammed Homan, Cecilia Söderberg-Nauclèr, Dept. of Medicine, Karolinska Institute, Stockholm
Abstract: Human Cytomegalo Virus (HCMV) is a rather unexplored virus belonging to the herpes virus family. The goal of this project is to segment, classify, and describe virus capsids at different maturation stages from transmission electron microscopy (TEM) images of infected cell nuclei. The virus capsids are to be classified to three different stages of the maturation pathway. The segmentation is done by template matching for one class at a time. The templates were produced from normalised radially averaged images of a number of typical particles of each class. A journal paper describing the method was published in Computer Methods and Programs in Biomedicine in 2004. During the year a means of measuring certain structural features of the capsids was also developed.
Carolina Wählby, Joakim Lindblad, Ewert Bengtsson
Funding: Amersham Biosciences, Uppsala, Cardiff, UK; UU TN-faculty; (This project was previously part of the Swedish Foundation for Strategic Research VISIT programme)
Period: 9902-0401
Partners: Lennart Björkesten, Amersham Biosciences, Uppsala; Stuart Swinburne, Simon Port, Alla Zaltsman, Gareth Bray, and Dietrich Ruehlmann, Amersham Biosciences, Cardiff, UK
Abstract: The interaction with and effect of potential drugs on living cells can be observed by fluorescence microscopy. High throughput methods for analysis of cells can be used as a tool in the drug discovery process. The overall objective of this project was to develop image analysis methods for segmentation, feature extraction, and classification of cells and sub-cellular structures in fluorescence microscopy images.
The cell nucleus has a well-defined shape and is relatively easy to detect. The cytoplasm is however more complex. The first goal of this project was to develop a fully automatic method for cytoplasm segmentation. The present algorithm, inspired by literature and previous experience, consists of an image pre-processing step, a general segmentation and merging step followed by a quality measure and a splitting step. By training the algorithm on one image, it is made fully automatic for subsequent images created under similar conditions. A summary of the results was presented in Pattern Recognition and Image Analysis in 2004.
Based on the experience from the segmentation of cytoplasms, a more problem specific project was initiated in cooperation with Amersham Biosciences in Cardiff, UK, in 2002. The aim of this project was to develop algorithms enabling fully automatic, real-time segmentation and analysis of fluorescence images of cells so as to quantitatively estimate the IGF-1 induced translocation of GFP-Rac1 to the cellular membrane for individual cells. Due to the ultimate goal of analyzing one image containing roughly 200 cells in less than two seconds, effort was taken not to use algorithms of high time complexity. The results were presented in Cytometry in January 2004.
Patrick Karlsson, Joakim Lindblad, Carolina Wählby, Ewert Bengtsson
Funding: UU TN-faculty; (This project was previously part of the Swedish Foundation for Strategic Research VISIT programme)
Period: 0305-
Partners: Mats Nilsson, Department of Genetics and Pathology, Uppsala University
Abstract: The interior of a cell is elaborately subdivided into many functionally distinct compartments, often organized into intricate systems. One way of studying such compartments is by the use of different fluorescent markers that bind specifically to the objects of interest. This type of staining followed by imaging through a microscope often results in point-source signals, or "blobs", together with a background of noise and autofluorescence. Analysis of spatial relationships requires pre-processing followed by separation and segmentation of the different blobs by combining intensity and shape information. Once the different blobs are detected, the goal is to detect non-random patterns in the blob distribution. True patterns were compared with synthetic model patterns created by hypothesis-based placement of blobs. Part of the work presented at an international conference (ISBI-04).
Carolina Wählby, Amalka Pinidiyaarachchi, Magnus Gedda, Patrick Karlsson, Ewert Bengtsson
Funding: Swedish Research Council; UU TN-faculty; (This project was previously part of the Swedish Foundation for Strategic Research VISIT programme)
Period: 0306-
Partners: Karin Althoff, Johan Degerman, Tomas Gustavsson, Jonas Faijerson, Torsten Olsson and Peter Eriksson, Chalmers University of Technology, Göteborg, Dept. of Clinical Neuroscience, Göteborg University
Abstract: Stem cells are cells that have the capacity to both renew themselves and generate progeny of more than one type. The field of stem cell research has rapidly evolved as a promising area in medicine, and one way of verifying the characteristics of stem cells is by time-lapse microscopy of cells in culture. Our partners at Chalmers have developed a time-lapse microscopy system equipped with a computer controllable motorized stage for automated compensation of stage motion displacement and auto focus. The system is suitable for in-vitro stem cell studies and allows for multiple cell culture image sequence acquisition, tracking and migration analysis. The goal of this project is to develop automated segmentation and tracking methods for comparative studies concerning rate of cell splits, cell motion analysis as a function of progeny type. Stable and robust tracking methods require a combination of segmentation and feature extraction combined with multiple hypothesis testing for tracking. Segmentation results based on variance filtering and seeded watershed segmentation are shown in Figure 4. A guest student Tang Chunming, from Harbin University, China, is also taking part in this project.
|
Carolina Wählby, Ida-Maria Sintorn, Ewert Bengtsson, Gunilla Borgefors
Funding: UU TN-faculty; SLU S-faculty; (This project was previously part of the Swedish Foundation for Strategic Research VISIT programme)
Period: 0209-
Partners: Fredrik Erlandsson, Dept. of Oncology/Pathology, Karolinska Institute, Stockholm
Abstract: Shape and distribution of various sub-cellular structures and components can be observed by immunostaining and in situ hybridization of fluorescent markers followed by fluorescence microscopy. The 3D images are acquired by making non-invasive serial optical sections of the object. Studies of the distribution of signaling factors involved in the cell cycle control indicate that minor changes in the signaling systems are the first signs of cancer transformation and tumor formation. Understanding the 3D organization of normal and transformed cell nuclei is therefore of great interest as a new approach to understanding the pathways of cancer. All image cytometry requires robust segmentation techniques. Clustered objects, background variation, as well as internal intensity variations complicate the segmentation of cells in tissue. An algorithm for segmentation of images of cell nuclei in tissue that combines intensity, shape, and gradient information has been developed and tested on 2D as well as 3D data. The results were published in Journal of Microscopy in July 2004.
Xavier Tizon
Funding: SLU S-faculty; (This project was previously part of the Swedish Foundation for Strategic Research VISIT programme)
Period: 0301-0410
Partners: Örjan Smedby, Anders Persson, Adam Löfving, Dept. of Medicine and Care, Linköping University Hospital
Abstract: This project is aimed at selecting a subset of volumetric data, and presenting it in such a way as to make diagnosis easier. In magnetic resonance angiography (MRA), it is of great interest to be able to separate arteries from veins. This problem is not trivial, because the vessels can be close and parallel throughout the image, especially in the neck region. Our algorithm extends the concept of binary connectedness to grey-level connectedness, using fuzzy sets. As start sets, we used small sets of voxels marked by the user. Good user interaction possibilities, portability and reusability are important concerns in this project. That is why we provided an implementation as a ``plugin" for the public domain package ImageJ. Clinical validation is in progress at Linköping University Hospital.
Xavier Tizon, Gunilla Borgefors
Funding: SLU S-faculty; (This project was previously part of the Swedish Foundation for Strategic Research VISIT programme)
Period: 0209-0410
Partners: Håkan Ahlström, Hans Frimmel, Tomas Hansen, Lars Johansson, Joel Kullberg, Dept. of Oncology, Radiology, and Clinical Immunology, UU Hospital; Qingfen Lin, previously at Computer Vision Laboratory, Dept. of Electrical Engineering, Linköping University
Abstract: As part of a large clinical trial, launched in order to study multiple factors causing arteriosclerosis, a large number of volunteers are participating in a whole-body MR angiography study at UU Hospital. Our goal is to develop Image Analysis tools to derive global measures of arteriosclerosis, from the characteristics of a limited portion of the arterial tree. The volume image analysis is split in four sub-tasks:
(a) Registration of sub-volumes. The acquisition technique produces four MRA volumes that correspond to the head and upper torso, the abdomen, the upper legs, and the lower legs. The acquisition procedure is optimized for patient comfort, producing a whole-body data set in only a few minutes. As a consequence, images show considerable geometric distortion near the borders of the volume. We use a tubular phantom, consisting of plastic tubes filled with water and contrast media, to produce an image using the standard MRA protocol. This image is rectified, as the geometry of the object is known, to estimate the original distortion of MRA data. This work has also resulted in a Master Thesis Master thesis at the MR Department of UU Hospital.
(b) Segmentation and identification of the arterial structure. Once the full volume has been registered and reconstructed, the arteries are segmented from the rest of the visible structures. Using limited user interaction and a modified Fast Marching algorithm, we are able to identify a predetermined subset of the arterial tree, removing veins and arteries with low diagnostic interest. The proposed method computes a cost map, from which it is possible to find the best (i.e. lowest cost) path between any point in the image and a given starting point. A graph is built, representing a simplified version of the arterial tree, that can be used for comparison between patients, and for localization of detected features along the arteries.
(c) Geometric measures to give an estimate of plaque burden. The geometry of the tubular structure of the arterial tree has to be studied. Candidate measures indicating the plaque burden that have been developed are: irregularities in the diameter along the central line of the vessel; curvature of the vessels; torsion of the vessels. Differential geometrical methods are used, together with scale-space theory, to study the degree to which structures resemble tubes.
The potential measures must of course be clinically evaluated. testing. A clinical study will help to choose which ones are the best for the physician to use.
Pasha Razifar, Ewert Bengtsson
Period: 0110-
Fundig: Amersham Foundation, UU TN-Faculty
Partners: Mats Bergström, Harald Schneider, UU and Uppsala Imanet
Abstract: Positron emission tomography (PET) is a powerful imaging technique with the potential of obtaining functional or biochemical information by measuring distribution and kinetics of radiolabelled molecules in a biological system, both in vitro and in vivo. PET images can be used directly or after kinetic modeling to extract quantitative values of a desired physiological, biochemical or pharmacological entity. PET images are generally rather noisy, meaning that the individual images are not optimal for the analysis and visualization of anatomy and pathology. Therefore it is essential to understand how noise affects the derived quantitative values. A pre-requisite for this understanding is that the properties of noise such as variance (magnitude) and texture (correlation) are known.
In earliest phase of the project, a technique for studying the pattern of noise distributions and correlation in both synthetically and experimentally generated PET images, using autocor relation function (ACF) was developed. The results were illustrated as one-dimensional (1D) profiles and even visualized as two-dimensional (2D) ACF images, revealing information about the noise properties which was then further explored. Experimental PET data were acquired in 2D and 3D acquisition mode and reconstructed by both analytical filtered back-projection (FBP) and iterative ordered subsets expectation maximization (OSEM) methods. Also, the results from these studies were compared with results from covariance matrix. Furthermore ACF has been applied on other medical imaging modalities such as Single Photon Emission Computed Tomography (SPECT), Computed Tomography (CT) and PET-CT for investigation of noise properties in these imaging tools.
Average images have been used to reduce the noise in PET images, but these average images tend to dampen the differences between regions of different kinetics. Parametric images, aiming at extracting areas with specific kinetic properties can enhance the discrimination between regions and normal contra pathology, but such methods typically enhance noise or at least do not optimize signal-to-noise ratio. It is clear that in several types of PET studies, the existing methods for generating diagnostic images, using either summations or parametric images, are not optimal. Therefore, Principal component analysis (PCA) and Independent Component Analysis (ICA) have been studied on dynamic PET images to study if it is possible to generate images, which emphasized regions withdifferences in behavior.
Different types of normalization algorithms have been suggested and studied for better optimization of signal-to-noise ratio in PET images before applying PCA and ICA. These studies have even been followed by studies concerning application of these two multivariate methods on normalized PET images, utilizing time-activity data obtained by using a reference region. The methods are adopted for certain PET applications, notably for the diagnosis of Alzheimers disease with a new amyloid binding PET tracer. The methods are probed and fine tuned both on synthetic and clinical PET images with the purpose to optimize the signal to noise ratio in these images.
![\includegraphics[width=0.27\textwidth]{Images/Xa.ps}](img24.png)
![\includegraphics[width=0.27\textwidth]{Images/Xb.ps}](img25.png)
![\includegraphics[width=0.27\textwidth]{Images/Xc.ps}](img26.png)