|
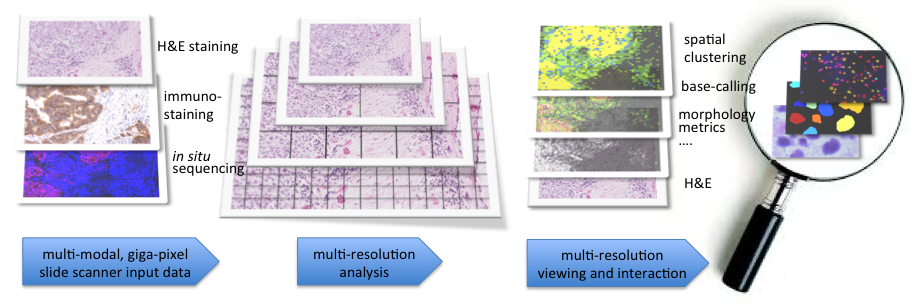
TissueMaps: Integrating spatial and genetic information via automated image analysis and interactive visualization of tissue data
Carolina Wählby, Petter Ranefall, Omer Ishaq, Marco Mignardi
Partners: Mats Nilsson, Thomas Hauling, Xiaoyan Qian, Jessica Svedlund, Elin Lundin, SciLifeLab/Stockholm University, Ola Söderberg and Gaëlle Cane, SciLifeLab/Uppsala University
Funding: Science for Life Laboratory; TN-faculty, UU; International Postdoctoral fellowship to Marco Mignardi
Period: 1109-
Abstract: Digital imaging of tissue samples and genetic analysis by next generation sequencing are two rapidly emerging fields in pathology. The exponential growth in digital imaging in pathology is catalyzed by more advanced imaging hardware, comparable to the complete shift from analog to digital images that took place in radiology a couple of decades ago: Entire glass slides can be digitized at near the optical resolution limits in only a few minutes' time, and fluorescence as well as bright field stains can be imaged in parallel.
Genetic analysis, and particularly transcriptomics, is rapidly evolving thanks to the impressive development of next generation sequencing technologies, enabling genome-wide single-cell analysis of DNA and RNA in thousands of cells at constantly decreasing costs. However, most of today's available technologies result in a genetic analysis that is decoupled from the morphological and spatial information of the original tissue sample, while many important questions in tumor- and developmental biology require single cell spatial resolution to understand tissue heterogeneity.
In this project, we develop computational methods that bridge these two emerging fields. We combine spatially resolved high-throughput genomics analysis of tissue sections with digital image analysis of tissue morphology. Together with collaborators from the biomedical field, we work with advanced digital image processing methods for spatially resolved genomics (see Ke et al, Nature Methods 2013). Going beyond visual assessment of this rich digital data will be a fundamental component for the future development of histopathology, both as a diagnostic tool and as a research field. We published a review paper on spatially resolved genomics and proteomics in the Journal of Molecular Biology (Koos et al J Mol Bio 2015).

|
Evaluation of the Effect of Compaction Oligonucleotides on the Strength and Integrity of Fluorescent Signals
Omer Ishaq, Petter Ranefall, Carolina Wählby
Partners: Carl-Magnus Clausson, Linda Arngården, Ola Söderberg, Dept. of Immunology, Genetics and Pathology, UU
Funding: Science for Life Laboratory
Period: 1310-1506
Abstract: Rolling circle amplification (RCA) performs nucleic acid replication for rapid synthesis of multiple concatenated copies of circular DNA. These molecules can be visually observed through the use of fluorescent markers. Moreover, the introduction of a compaction oligonucleotide during RCA results in brighter and more compact signals. The project aims to evaluate the effect of compaction oligonucleotides on the strength and integrity of fluorescent signals. A paper describing the method, including image analysis approaches for methods evaluation, was published in Nature Scientific Reports 2015.
Detection of Fluorescent Signals using Deep Learning Architectures
Omer Ishaq, Carolina Wählby
Partners: Vladimir Curic, Martin Linden, Johan Elf, Dept. of Cell & Molecular Biology, UU
Funding: Science for Life Laboratory, eSSENCE, VR junior researcher grant to Carolina Wählby
Period: 1501-
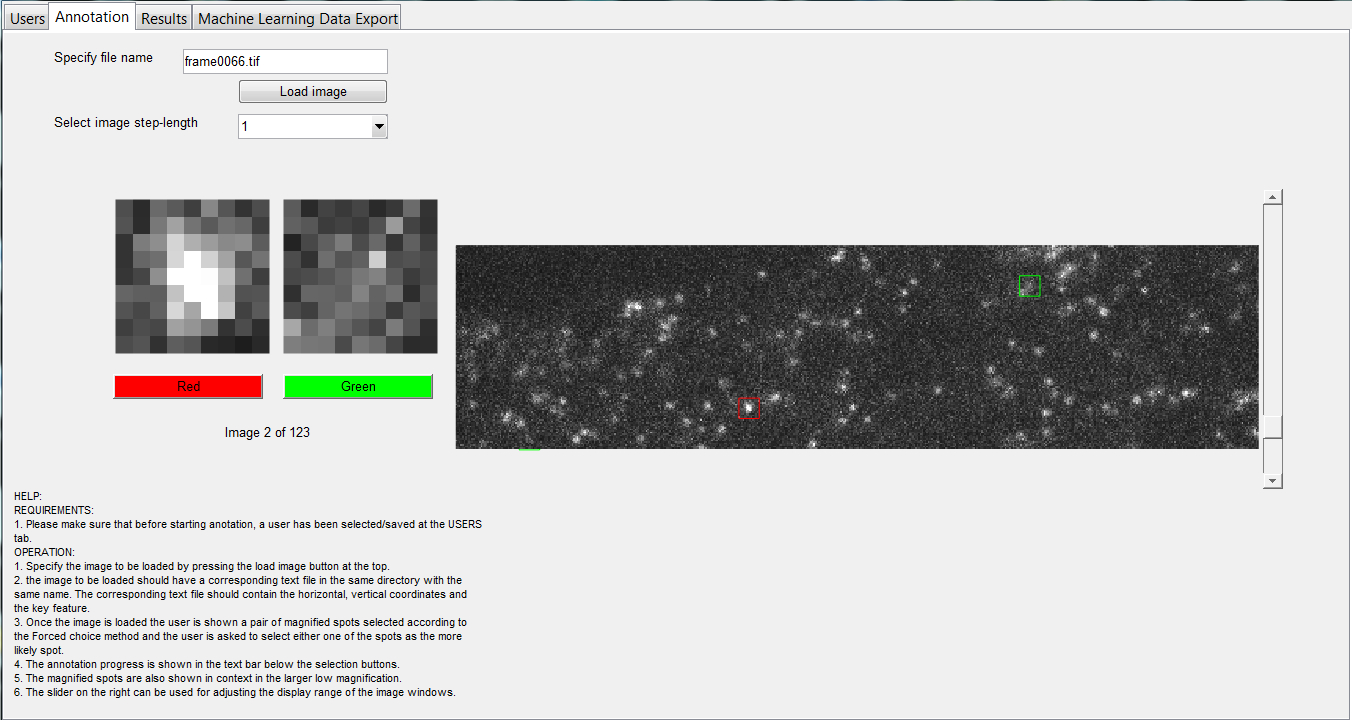
Abstract: Detection of fluorescent spots is an important component of bioimaging. A number of detection methods have been proposed. Recently, deep learning methods have become popular for a range of computer vision tasks and have resulted in competitive results. In this project we utilize a number of these deep learning methods and compare them against model-based spot detection methods. In addition, we also explore the effect of training both shallow- and deep-learning spot detection approaches on synthetic, semi-synthetic and real data and evaluate their performance on manually annotated real data in the form of quantitative results. The annotation of real data is facilitated by the development of a specialized annotation tool based on a two-alternative forced-choice (2AFC) approach. The annotation performance is validated through rater reliability statistics. The project has resulted in two manuscripts, one of which has been submitted to a conference and the other is being adapted for a journal publication.
Figure 12 shows the interface of the two-alternative forced-choice (2AFC) tool developed for spot annotation.
|
SciLifeLab Cancer Stem Cell Program
Damian Matuszewski, Petter Ranefall, Carolina Wählby, Ida-Maria Sintorn
Partners: Sven Nelander, Ingrid Lönnstedt, Cecilia Krona, Linnèa Schmidt, Karin Forsberg-Nilsson, Irina Alafuzoff, Ulf Landegren, Anna Segerman, Tobias Sjöblom, Lene Urborn, and Bengt Westermark, Dept. of Immunology, Genetics and Pathology and SciLifeLab, UU, Bo Lundgres, the Karolinska Institute and SciLifeLab, Stockholm, Rebecka Jörnsten, Chalmers, Gothenburg, and Göran Hesselager, UU Hospital, Uppsala
Funding: AstraZeneca-Science for Life Laboratory Joint Research Program
Period: 1303-
Abstract: The SciLifeLab Cancer Stem Cell Program is a cross-platform initiative to characterize cancer stem cells (CSCs). Previously, the development of drugs targeting the CSC population in solid tumors has been curbed by the lack of valid cell model systems, and the complex genetic heterogeneity across tumors, factors that make it hard to assess new targets or predict drug responses in the individual patient. To solve these problems, our aim is to develop a biobank of highly characterized CSC cultures as a valid model of cancer heterogeneity. We will combine mathematical and experimental approaches, including image-based high-throughput cell screening, to define the spectrum of therapeutically relevant regulatory differences between patients. This will help elucidate mechanisms of action and enable accurate targeting of disease subgroups. Patient data is continuously collected, and close to one hundred primary cell lines have been established. The cultured cells are exposed to known and novel drug compounds at varying doses, and imaged by fluorescence as well as bright-field microscopy. In 2015 algorithms for cell cycle analysis and automatic selection of potentially effective treatments were developed, and presented at BioImage Informatics 2015 in Gaithersburg, MD, USA. Current research focus is on extracting meaningful morphological descriptors from the image data. As part of the project we also evaluate the infiltration of tumor cells upon injection of stem cells in mice brains.
PopulationProfiler
Damian Matuszewski, Carolina Wählby, Ida-Maria Sintorn
Partners: Jordi Carreras Puigvert, SciLifeLab and Helleday Laboratory, Karolinska Institutet, Stockholm
Funding: Science for Life Laboratory
Period: 1501-
Abstract: PopulationProfiler is a cross-platform open-source tool developed for data analysis in image-based screening experiments. The main idea is to reduce per-cell measurements to per-well distributions, each represented by a histogram. These can be optionally further reduced to sub-type counts based on gating (setting bin ranges) of known control distributions and local adjustments to histogram shape. Such analysis is necessary in a wide variety of applications, e.g. DNA damage assessment using foci intensity distributions, assessment of cell type specific markers, and cell cycle analysis.
The source code, sample dataset and an executable program (for Windows only) are freely available at http://www.cb.uu.se/~damian/PopulationProfiler.html.
Segmentation and Tracking of E.coli Bacteria in Bright-Field Microscopy Images
Global and Local Adaptive Gray-level Thresholding Based on Object Features
Petter Ranefall, Sajith Kecheril Sadanandan, Carolina Wählby
Funding: Science for Life Laboratory
Period: 1501-1512
Abstract: We have developed two new algorithms for global and local adaptive thresholding based on object features such as size, ellipse fit, etc. The algorithms are efficient with little computational overhead compared to histogram based gray-level thresholding, but with much more stable results. This makes them very suitable for high-throughput analysis in microscopy applications like segmentation of cell nuclei or fluorescent spots. The algorithms have been implemented as plugins to ImageJ and CellProfiler and have been used in several different applications. We have written two papers that both are accepted for publication in Cytomerty A and at ISBI 2016.
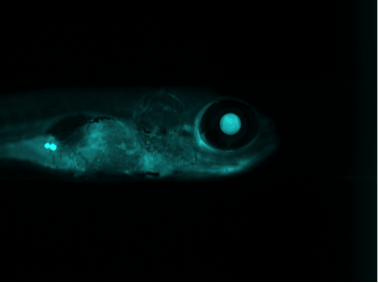
Quantification of Zebrafish Lipid Droplets
Petter Ranefall, Carolina Wählby
Partners: Marcel den Hoed, Manoj Bandaru, Erik Ingelsson, Dept. of Medical Sciences and SciLifeLab, UU
Funding: Science for Life Laboratory
Period: 1308-
Abstract: The aim of this project is to identify novel targets for the therapeutic intervention of coronary artery disease. This is done by following-up results from genome-wide association studies in epidemiological studies using a zebrafish model system. Using image analysis we try to identify and characterize causal genes within loci that have so far been identified as associated with coronary heart disease by (high-throughput) screening of atherogenic processes in wildtype and mutant zebrafish, both before and after feeding on a control diet or a diet high in cholesterol. Using confocal microscopy we can image fat accumulation in the zebrafish. We have also developed methods for length and volume measurements as well as quantification of macrophages, neutrophils, IK17 and the overlap with these expressions and stationary lipids. Our results confirm that zebrafish larvae represent a promising model system for early-stage atherosclerosis.
|
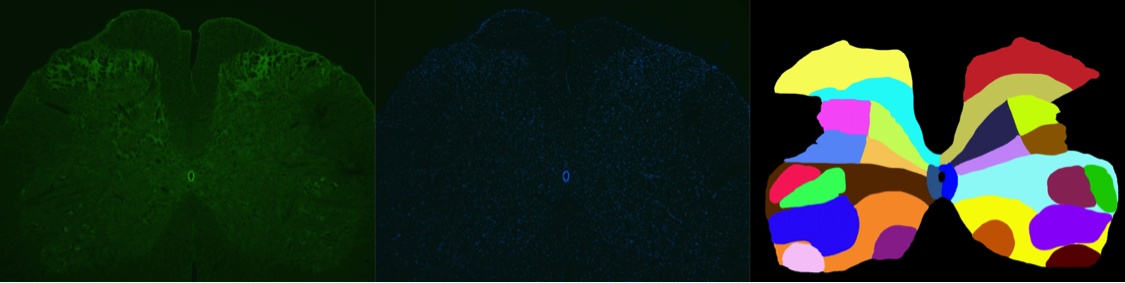
Rat Spinal Cord
Petter Ranefall, Carolina Wählby
Partners: Lada Stålhandske, Wei Sun, Georgy Bakalkin, Dept Pharm Biosci, UU
Funding: Science for Life Laboratory
Period: 1506-
Abstract: Our collaborators newly established an way of staining the activity in the endogenous opioid system in the rat spinal cord, and the aim of the project is to quantify the ammount and localization of mRNA staining. We have developed image analysis approaches for quantifying the amount of cells with positive signals and associate those to manually outlined regions of interest within the spinal cord of rats. We have applied our new method for local adaptive thresholding based on ellipse fit to segment nuclei, and use ilastik to classify positive/negative cells.
|
Infiltration of T Cells in Thyroid Glands
Petter Ranefall, Carolina Wählby
Partners: Susanne Kerje, Dept of Medical Biochemistry and Microbiology, UU
Funding: Science for Life Laboratory
Period: 1504-
Abstract: The aim of the project is to estimate the degree of infiltration of T cells in thyroid glands of chicken in order to better understand auto-immunity and rare genetic disease. We have developed an image analysis pipeline, using ilastik, CellProfiler, and some Python scripts, that extracts the regions of interest from the full glass slide images and classifies the tissue into infiltration or normal. The whole dataset contained 558 slides and the analysis was run on a powerful local server at the BMC.
Ubiquitin Screen
Petter Ranefall, Carolina Wählby
Partners: Johan Boström, Jordi Carreras Puigvert, Mikael Altun, Molecular Biochemistry and Biophysics, KI
Funding: Science for Life Laboratory
Period: 1502-
Abstract: Ubiquitin is a small protein that is found in almost all cellular tissues in humans and other eukaryotic organisms, which helps to regulate the processes of other proteins in the body. Cultured cells respond to treatments such as silencing of genes or exposure to radiation and/or drugs by changing their morphology, giving us hints on mechanisms of action. We develop methods for image-based high-throughput screening to identify subtle changes in individual cells, not accessible by bulk-methods, here focusing on the ubiquitin pathway.
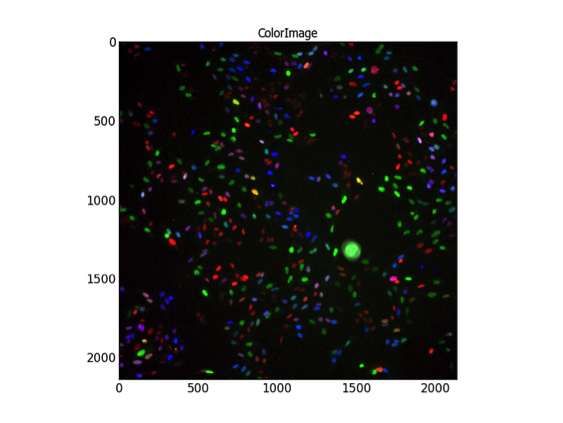
|
Cell Distribution and Protein Expression in the Ectocervix
Petter Ranefall, Carolina Wählby
Partners: Anna Gibbs, Maria Röhl, Annelie Tjernlund, Dept. of Medicine, KI
Funding: Science for Life Laboratory
Period: 1504-
Abstract: This research project is focused on mucosal immunology in the female genital tract and HIV. The female genital mucosa presents a comprehensive natural immune defense against HIV infection, although during exposure to a high dose of virus this is not enough to protect the individual against viral transmission. Some individuals have a stronger resistance against HIV than others and therefore it is highly important to investigate which factors that contribute to an effective local protection against sexual infection. The aim of this study is to quantify gene expression in the target cells of HIV in ectocervix, and measure the distance to the vaginal lumen, as well as epithelial thickness. These parameters will be compared in women involved in sex work between the groups of HIV-infected, highly HIV exposed HIV uninfected that seems to be resistant, and HIV uninfected women who have been involved in sex work for a short period.
Segmentation of Neurons
Petter Ranefall, Carolina Wählby
Partners: Laureanne Pilar Lorenzo, Niklas Dahl, Dept of Immunology, Genetics and Pathology, UU
Funding: Science for Life Laboratory
Period: 1402-
Abstract: The goal of this project is to analyze neurons grown from stem cells in vitro. The aim is to assess the percentage of neurons (using B-tubulin) and certain neuron subtypes (GABA) by immunofluorescence. We used CellProfiler to segment the cells and CellProfiler Analyst to classify positive cells.
Cell Time-Lapse Analysis
Petter Ranefall, Carolina Wählby
Partners: Grigorios Kyriatzis, Jennifer Feenstra, Theresa Vincent, Physiology and Pharmacology, KI
Funding: Science for Life Laboratory
Period: 1410-
Abstract: The aim of the project is to interpret differences in migration-proliferation of cells with different treatments and express those in a quantitative manner. We used a 'scratch assay' approach, or 'wound healing assay' as it sometimes is called, where cells are grown in wells, and then the surface is 'scratched' and loose cells are washed away. Then the wells are imaged, possibly followed by adding a drug substance, and imaging the wells again at a suitable time interval. The area filled at time point T is a measure of the migration speed.
Kidney Morphology and Topology of the Glomerular Filtration Barrier
Petter Ranefall, Carolina Wählby
Partners: David Unnersjö Jess, Hans Gunnar Blom, Dept of Applied Physics, KTH
Funding: Science for Life Laboratory
Period: 1510-
Abstract: Our collaborators have developed a super-resolution immunofluorescence microscopy protocol for the study of the filtration barrier in the kidney. The aim of the project is to quantitatively evaluate the morphology and topology of the glomerular filtration barrier in the kidney. The most promising approach for analyzing these challenging 3D images seems to be a 3D version of the adaptive local thresholding based on ellipse fit.
|
Vascular Networks
Petter Ranefall, Carolina Wählby
Partners: Elisabet Olin, Ross Smith, Chiara Testini, Lena Claesson-Welsh, Dept of Immunology, Genetics and Pathology, UU
Funding: Science for Life Laboratory
Period: 1406-
Abstract: In this project we analyze vascular networks in the mouse brain, retina networks and cell junction activations. We have several applications where we skeletonize the networks and extract branch points in the skeleton. For the cell junction activations we have initially used an approach where we compute the area of the activated junctions (green) between the cells and use that as a measurement of activation.
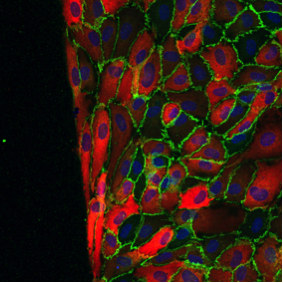
|
Objective Automated Quantification of Fluorescence Labeling in Histologic Sections of Rat Lens
Carolina Wählby, Nanna Zhou Hagström
Partners: Per Söderberg and Nooshin Talebizadeh, Dept. of Neuroscience, UU
Funding: Science for Life Laboratory
Period: 1501-
Abstract: The lens epithelium of the eye is a single layer of cells covering the anterior face of the lens. In this project we study how UV light affects the lens epithelial cells by quantitatively analyzing fluorescent signal from biomarkers in cell nuclei and cytoplasms. We have developed an automated method to delineate lens epithelial cells and to quantify expression of fluorescent signal of biomarkers in each nucleus and cytoplasm of lens epithelial cells in a histological section.
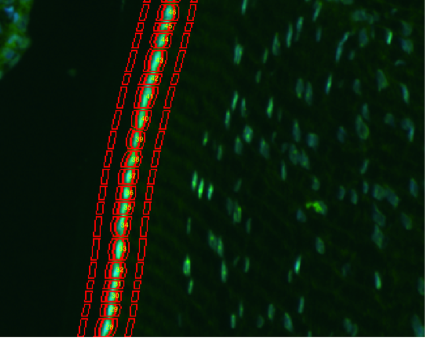
|
A Model System for Analysis of Spinal Cord Injury
Carolina Wählby
Partners: Nils Hailer and Nikos Schizas, Dept. of Surgical Sciences, UU
Funding: Science for Life Laboratory
Period: 1501-
Abstract: Following spinal cord injury neurons die due to neurotoxicity and inflammation. We study these effects in a model system with spinal cord slice cultures, aiming to find methods to reduce neurotoxicity. Our focus is quantitative image analysis methods that delineate activated cells and quantify protein expression as a response to injury and treatment.
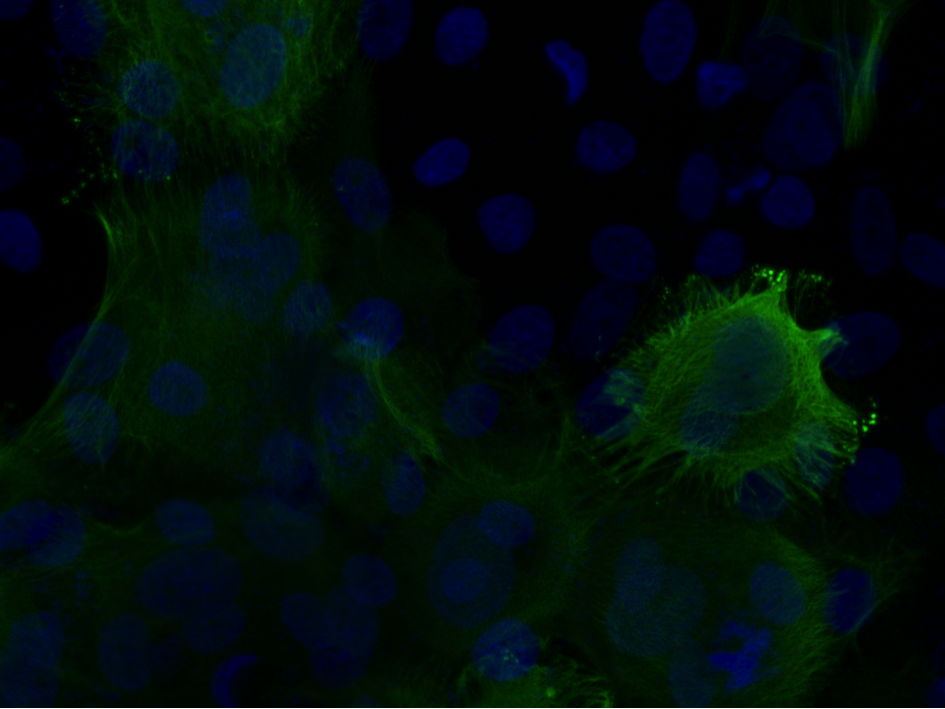
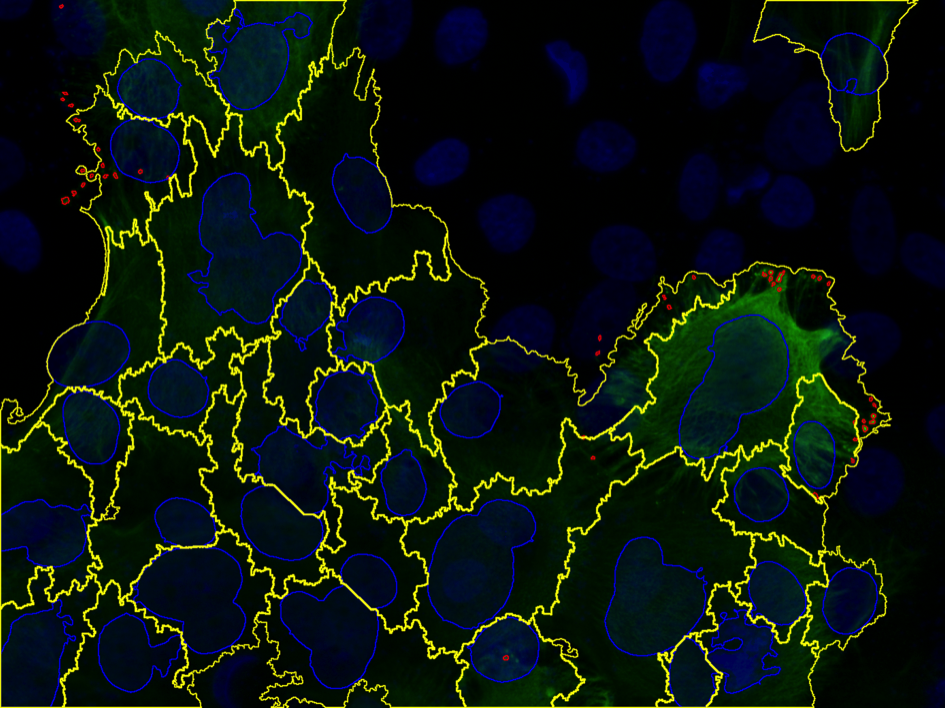
Analysis of Keratin Aggregates
Petter Ranefall, Carolina Wählby
Partners: Hanqian Zhang and Hans Törmä, Dept. of Medical Sciences, Dermatology and Venereology
Funding: Science for Life Laboratory
Period: 1510-
Abstract: Epidermolytic hyperkeratosis (EH) is a rare genetic skin disorder caused by mutation of keratin 1 or 10, and characterized by blistering in the epidermis and hyperkeratosis. The skin may blister easily following mechanical injury and exposure to heat etc. Immortalized keratinocyte cell lines were established by our collaborators at the Dept. of Medical Sciences, Dermatology and Venereology, and these cell lines show promise as a screening model to test new potential drugs for treating EH patients. Large-scale screening requires robust, efficient and effective image analysis methods, and we are currently developing methods to analyze keratin aggregates in cultured EH cells.
|