Ingrid Carlbom, Pontus Olsson, Fredrik Nysjö, Johan Nysjö
Partners: Daniel Buchbinder (Icahn School of Medicine at Mount Sinai, New York, NY, USA); Stefan Johansson (Division of Microsystems Technology, Uppsala University and Teknovest AB); Andreas Thor, Dept. of Surgical Sciences, Oral & Maxillofacial Surgery, UU Hospital; Andres Rodriguez Lorenzo, Dept. of Surgical Sciences, Plastic Surgery, UU Hospital.
Funding: BIO-X ; Thuréus Stiftelsen
Period: 1501-1512
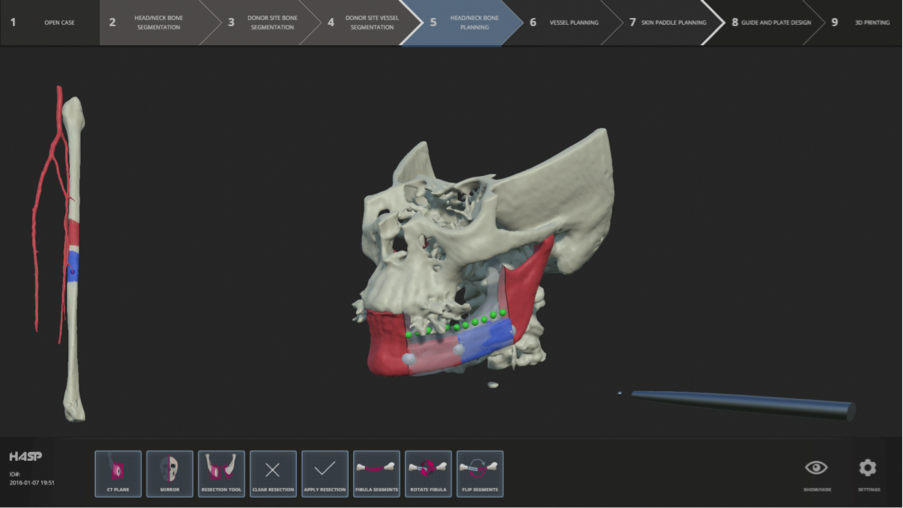
Abstract: This year our focus has been to build an integrated surgery planning system encompassing the entire planning process: from input of patient data to generation of saw guides and plates for the operating room. We employed a company, CHAS, which specializes in interface design. The result is a clear workflow, from data input, to bone segmentation, vessel segmentation, bone reconstruction, vessel reconstruction, soft tissue segmentation, and to the generation of guides and plates combined with descriptive icons representing the functions at each stage. We have transferred most of the old version of HASP into this new interface, and also begun to integrate BoneSplit (see below) for the bone segmentation.
In May, Pontus, Fredrik, and Ingrid spent a week in NYC at the Mount Sinai Beth Israel Hospital hosted by Dr. Buchbinder, where we demonstrated HASP and got feedback and input for improvements from about ten CMF surgeons, fellows, and residents.
In October, Pontus, Ingrid and her mentor Anders Lundqvist attended the yearly meeting of AAOMS, (American Association of Oral and Maxillofacial Surgeons) in Washington, DC, where Ingrid gave a talk on "Surgical Training Using a Haptics-Assisted Cranio-Maxillofacial Planning System (HASP)", and we demonstrated HASP to attendees.
In December, Pontus and Daniel Buchbinder demonstrated HASP at the AO Foundation Meeting in Davos, Switzerland. On October 16, Pontus Olsson successfully defended his thesis "Haptics with Applications to Cranio-Maxillofacial Surgery Planning", a large portion of which concerned HASP.
Ida-Maria Sintorn, Ingela Nyström, Fredrik Nysjö, Johan Nysjö, Anders Brun, Gunilla Borgefors
Partners: Dept. of Cell and Molecular Biology, Karolinska Institute; SenseGraphics AB, Dept. of Molecular Biology, Umeå University, Vironova AB
Funding: The Visualization Program by Knowledge Foundation; Vaardal Foundation; Foundation for Strategic Research; VINNOVA; Invest in Sweden Agency; SLU, faculty funding
Period: 0807-1412
Abstract: Electron tomography is the only microscopy technique that allows 3-D imaging of biological samples at nano-meter resolution. It thus enables studies of both the dynamics of proteins and individual macromolecular structures in tissue. However, the electron tomography images have a low signal-to-noise ratio, which makes image analysis methods an important tool in interpreting the images. The ProViz project aims at developing visualization and analysis methods in this area.
The development of the ProViz software ended 2014. The project activity in 2015 has been to prepare and finalize the manuscript describing and demonstrating the ProViz software in several different applications.
Robin Strand, Filip Malmberg
Partner: Joel Kullberg, Håkan Ahlström, Dept. of Radiology, Oncology and Radiation Science, UU
Funding: Faculty of Medicine, UU
Period: 1208-
Abstract: In this project, we mainly process magnetic resonance tomography (MR) images. MR images are very useful in clinical use and in medical research, e.g., for analyzing the composition of the human body. At the division of Radiology, UU, a huge amount of MR data, including whole body MR images is acquired for research on the connection between the composition of the human body and disease.
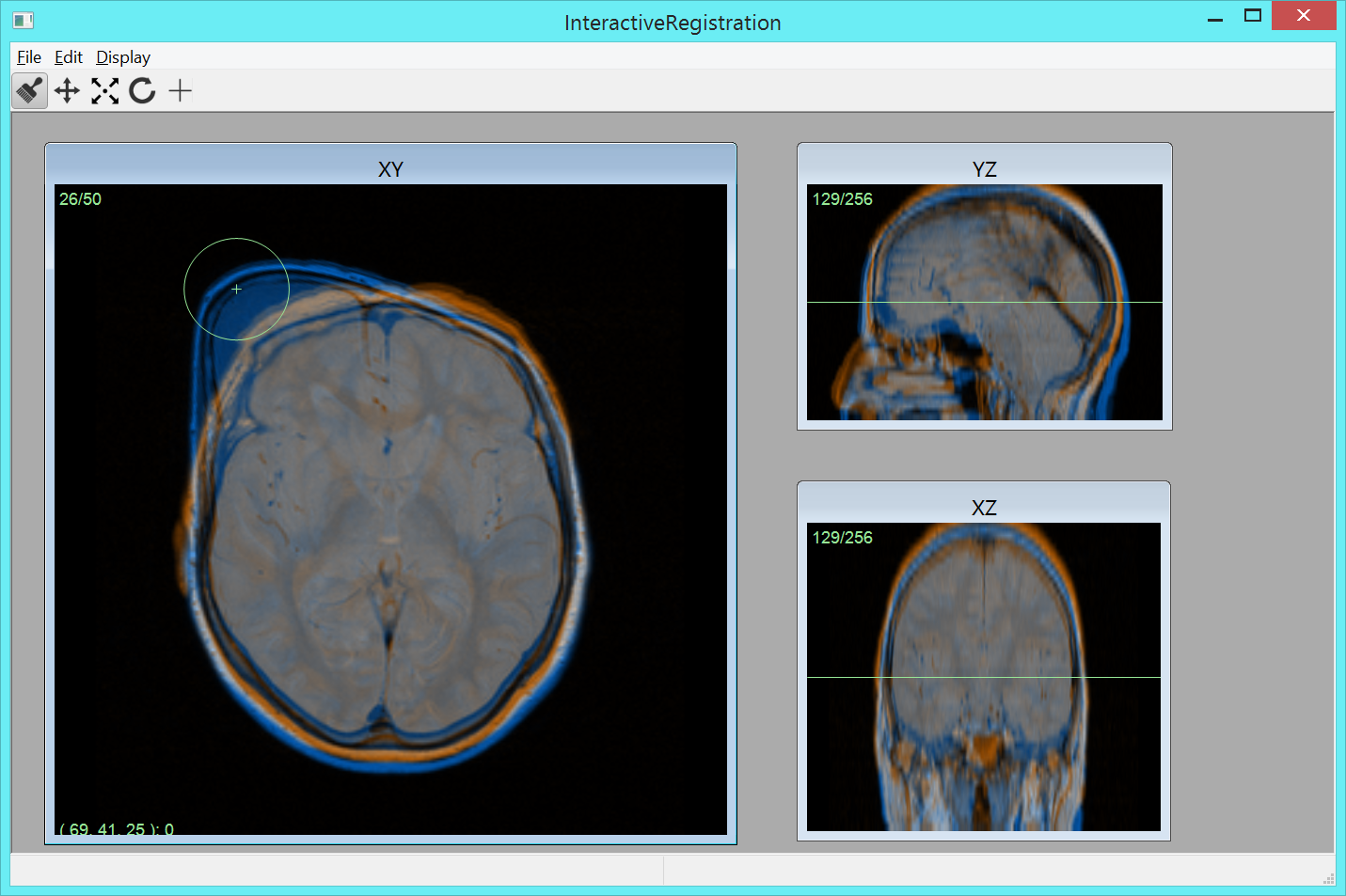
To compare volume images voxel by voxel, we develop image registration methods. For example, large scale analysis is enabled by image registration methods that utilizes, for example, segmented tissue (e.g., Project 6) and anatomical landmarks. Based on this idea, we have developed Imiomics (imaging omics) - an image analysis concept, including image registration, that allows statistical and holistic analysis of whole-body image data (Figure 5). The Imiomics concept is holistic in three respects: (i) The whole body is analyzed, (ii) All collected image data is used in the analysis and (iii) It allows integration of all other collected non-imaging patient information in the analysis. During 2015, we presented the Imiomics concept at the 23rd Annual meeting of the International Society for Magnetic Resonance in Medicine (MICCAI) in Toronto, Canada and at the Swedish conferences Medicinteknikdagarna (Medical Engineering Days) and Röntgenveckan (X-ray week). A manuscript describing a method for interactive manipulation of 3D deformation fields was presented at the Interactive Medical Image Computing (IMIC) workshop held in conjunction with the MICCAI conference.
|
Elisabeth Linnér, Robin Strand
Funding: TN-faculty, UU
Period: 1005-
Abstract: Three-dimensional images are widely used in, for example, health care. With optimal sampling lattices, the amount of data can be reduced by 20-30