Gustaf Kylberg, Ida-Maria Sintorn, Ewert Bengtsson, Gunilla Borgefors
Partners: Vironova AB; Delong Instruments, Brno, Czech Republic; Ali Mirazimi, Kjell-Olof Höglund, Centre for Microbiological Preparedness; Swedish Institute for Infectious Disease Control (SMI)
Funding: 2008-2011 Swedish Civil Contingencies Agency (MSB), Swedish Defense Materiel Administration (FMV), Swedish Agency for Innovative Systems (VINNOVA). 2011-2013 Eurostar project
Period: 0801-
Abstract: This project aims at automating the virus identification process in high resolution TEM images. This, in combination with Project 2 to create a rapid, objective, and user independent virus diagnostic system. The identification task consists of method development for segmenting virus particles with different shapes and sizes and extracting descriptive features of both shape and texture to enable the classification into virus species. Texture features such as variants of Local Binary Patterns and Regional Moments (filter banks constructed from orthogonal moments), are being evaluated on virus textures as well as other texture datasets to get a deeper understanding of the discriminant power of the features under different conditions. A paper comparing, combining and evaluating the discriminating power of local texture descriptors for virus classification was presented at the 22nd International Conference on Pattern Recognition (ICPR) in Stockholm in August and Gustaf Kylberg defended his PhD thesis closely linked to this project in March 2014.
Gustaf Kylberg, Ida-Maria Sintorn, Ewert Bengtsson, Gunilla Borgefors
Partners: Vironova AB; Delong Instruments, Brno, Czech Republic
Funding: Eurostar project
Period: 1107-
Abstract: Transmission electron microscopy (TEM) is an important clinical diagnostic and material analysis tool. Transmission electron microscopes are expensive, complex, sensitive and bulky machines, often housed in specially built rooms to avoid vibrations affecting the imaging process. They are to a very large extent manually operated, meaning that an expert in electron microscopy and preferably also in the application at hand needs to perform the analysis at the microscope, an often very time consuming task.
This project aimed at developing the MiniTEM, a desk-top low voltage TEM designed for imaging biological samples, with a high degree of automation regarding instrument alignment, image acquisition and analysis. The goal was a small, cheap, robust, and easy to use system that requires no more training than any simple lab equipment, and can be hosted in any office or lab (even mobile).
Automating the image acquisition process is key for reducing the manual input and making the imaging and analysis more objective. Within the project different strategies for automating the image acquisition were developed and investigated. The first was acquisition of images at random positions on the grid. The second was to search for a specific structure/object and only acquire (store) the images containing the structure/object of interest. The third was similar to the second approach but embedded in a multi-scale approach with the goal to make the acquisition more efficient.
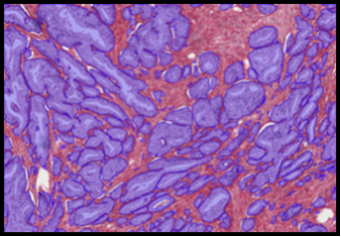
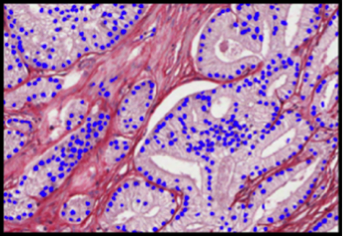
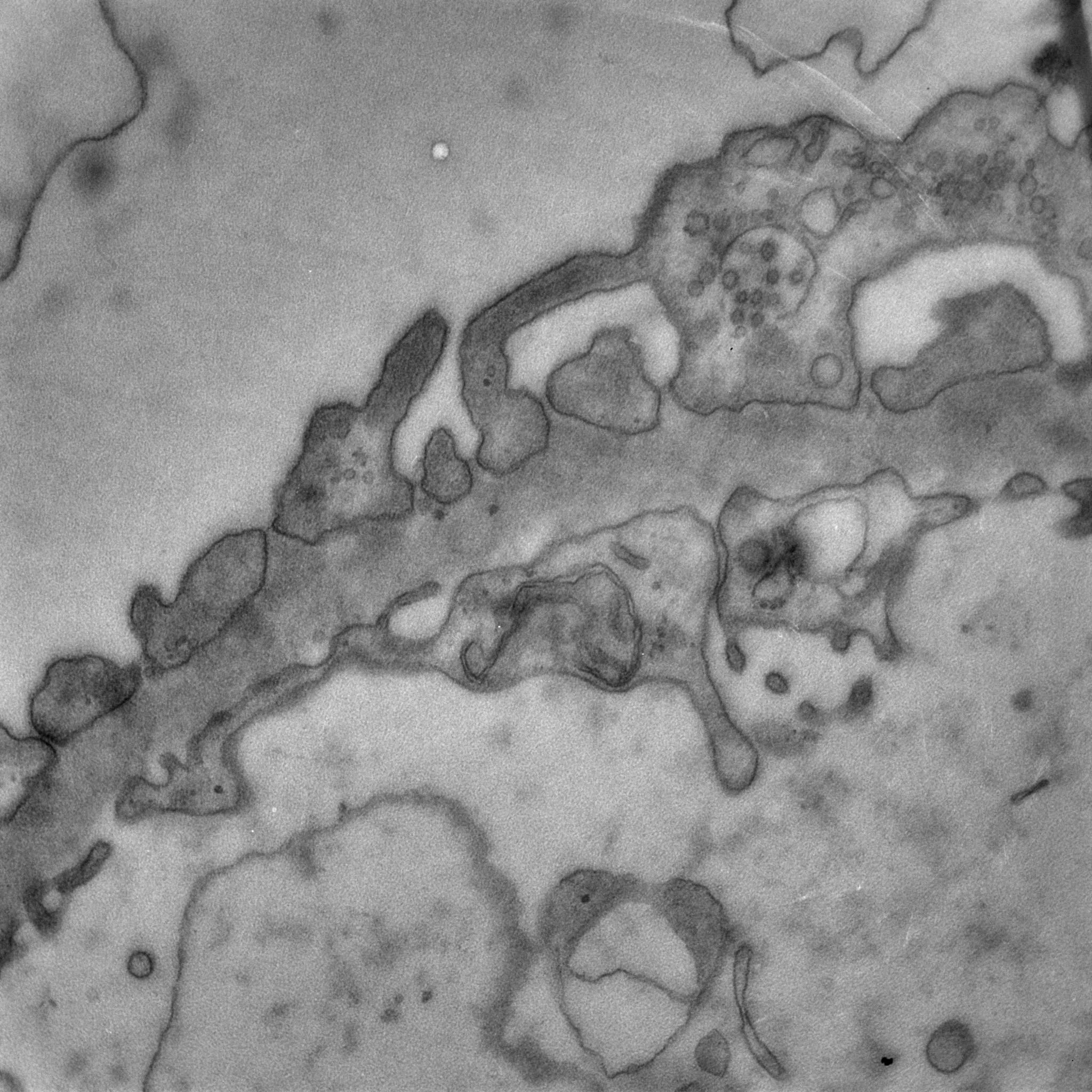
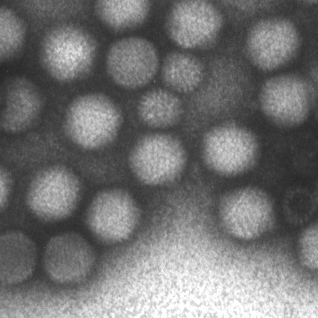
The MiniTEM was launched and displayed at the International Microscopy Congress in Prague in September. One instrument was at the conference site and a second piece, physically in Stockholm, was operated remotely from Prague. Abstracts and posters about the project, the developled image analysis methods, and actual instrument, have been presented at various international and national conferences and meetings throughout the year. Gustaf Kylberg successfully defended his PhD thesis early 2014, which was closely linked to this project. The MiniTEM, its GUI and two example images showing a mixture of typical clinical application areas are shown in Figure 3.
Omer Ishaq, Alexandra Pacureanu, Carolina Wählby
Partners: Johan Elf, Gustaf Ullman, Fredrik Persson, Dept. of Cell & Molecular Biology, UU
Funding: SciLifeLab Uppsala, eSSENCE, VR junior researcher grant to CW
Period: 1211-
Abstract: Stochastic optical reconstruction microscopy (STORM) is a super-resolution microscopy image acquisition technique for single-molecule localization. Like other stochastic super-resolution microscopy techniques it incorporates a trade-off between spatial- and temporal-resolution. Recently, a compressed-sensing (CS) based variant of STORM, called FasterSTORM, has been developed which substantially increases the temporal sampling of a stack of STORM image frames. This improvement is realized by increasing the density of activated fluorophores in each frame, followed by a subsequent CS-based retrieval of single-molecule positions even with overlapping fluorescent signals. However, the CS-based retrieval/decoding step is time consuming and can take as much as three hours for each image frame. We have accelerated the FasterSTORM method through parallel processing on multi-core processors. Additionally, we have tested and tried a number of L1-solvers for CS-based recovery of molecule positions. We are in the process of comparing the performance of the FasterSTORM against a wavelet-based approach to localize fluorescent signals in time-lapse images of bacterial cells.
In 2014, a paper comparing convex and greedy solvers and evaluating the sensitivity of the FasterSTORM to estimation bias of the point spread function (PSF) was accepted for oral presentation at the 22nd International Conference on Pattern Recognition (ICPR 2014).
|

|
Carolina Wählby, Alexandra Pacureanu, Petter Ranefall
Partners: Mats Nilsson, Rongqin Ke, Marco Mignardi, Thomas Hauling, Xiaoyan Qian, SciLifeLab Stockholm/Stockholm University
Funding: SciLifeLab Uppsala; TN-faculty, UU
Period: 1109-
Abstract: Profiling of gene expression is prerequisite for understanding the function of cells, organs and organisms, in health and disease. The sequencing techniques currently in use rely on homogenization of the samples. Therefore, the obtained information represents either the average expression profile of the tissue sample or expression profiles of isolated single cells. Our collaborators have developed a new molecular method, enabling in situ sequencing of mRNA, so that protein expression can be observed directly in cultured cells or tissue samples. We have developed image analysis tools for automated analysis of sequencing data, mapping, and visualization of gene expression patterns (Figure 4). We presented the work as part of a Special Session on Advances in Computer-Aided Histopathology at the IEEE International Symposium on Biomedical Imaging (ISBI 2014) in Beijing. The project was also presented at the 1st annual conference for the Society of Biomolecular Imaging and Informatics SBI2, at the JB Martin Conference Center at Harvard Medical School, Boston, MA, USA, where Carolina Wählby was honored with the SBI2 `President's innovation award' for her presentation on `Combining image-based in situ RNA screening with quantitative analysis of cell and tissue morphology'.
|
Omer Ishaq, Petter Ranefall, Carolina Wählby
Partners: Carl-Magnus Clausson, Linda Andersson, Ola Söderberg, Dept. of Immunology, Genetics and Pathology, UU
Funding: SciLife Lab Uppsala
Period: 1310-
Abstract: Rolling circle amplification (RCA) performs nucleic acid replication for rapid synthesis of multiple concatenated copies of circular DNA. These molecules can be visually observed through the use of florescent markers. Moreover, the introduction of a compaction oligonucleotide during RCA results in brighter and more compact signals. The project aims to evaluate the effect of compaction oligonucleotides on the strength and integrity of florescent signals. A manuscript has been submitted for journal publication.
Damian Matuszewski, Carolina Wählby, Ida-Maria Sintorn
Partners: Sven Nelander, Karin Forsberg-Nilsson, Irina Alafuzoff, Ulf Landegren, Anna Segerman, Tobias Sjöblom, Lene Urborn and Bengt Westermark, Dept. of Immunology, Genetics and Pathology and SciLifeLab, UU, Bo Lundgren, the Karolinska Institute and SciLifeLab, Stockholm, Rebecka Jörnsten, Chalmers, Gothenburg, and Göran Hesselager, UU Hospital, Uppsala
Funding: AstraZeneca-Science for Life Laboratory Joint Research Program
Period: 1303-
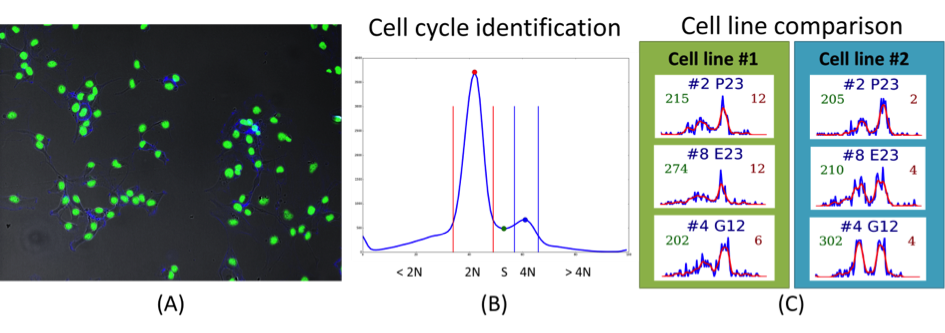
Abstract: The SciLifeLab Cancer Stem Cell Program is a cross-platform initiative to characterize cancer stem cells (CSCs). Previously, the development of drugs targeting the CSC population in solid tumors has been curbed by the lack of valid cell model systems, and the complex genetic heterogeneity across tumors, factors that make it hard to assess new targets or predict drug responses in the individual patient. To solve these problems, our aim is to develop a biobank of highly characterized CSC cultures as a valid model of cancer heterogeneity. We will combine mathematical and experimental approaches, including image-based high-throughput cell screening, to define the spectrum of therapeutically relevant regulatory differences between patients. This will help elucidate mechanisms of action and enable accurate targeting of disease subgroups. During 2013-2014, patient data was collected, and a number of primary cell lines were established. Cultured cells were exposed to a different treatments and doses (more than 2500 different treatments per cell line), and imaged by fluorescence as well as bright-field microscopy, and current focus is on extracting meaningful morphological descriptors from the image data (Figure 5). Developed tools are also applied in a cell cycle analysis project realized together with Jordi Carreras Puigvert, the Karolinska Institute and SciLifeLab, Stockholm.
|
Bettina Selig, Cris Luengo
Partners: Bernd Rieger, Quantitative Imaging Group, Delft University of Technology, The Netherlands; Koen Vermeer, The Rotterdam Eye Hospital, The Netherlands
Funding: S-faculty, SLU
Period: 1103-1412
Abstract: The corneal endothelium plays a key role in maintaining the transparency of the cornea.Because the cells in the endothelium do not regenerate, the cell density decreases with age; this reduces its ability to maintain the processes needed to keep the cornea transparent. Thus, being able to measure this density in patients is very important. The endothelium can be imaged by specular microscopy or by confocal scanners, and measurements can be obtained manually, automatically with manual corrections, or fully automatically with current software (e.g., Nidek's NAVIS). Unfortunately, the results of the automatic methods are often useless, especially at low cell densities. Together with the Rotterdam Eye Hospital, we have developed a fully automatic method to segment individual cells in the corneal endothelium. The result of the method can be used to determine the cell density, but also other parameters of interest, like pleomorphism (cell shape) and polymegathism (cell size variation). Our segmentation method produces a segmentation that matches a manual segmentation reasonably well, for a wide range of cell densities and image qualities. These results have been submitted for publication during 2014.
Ewert Bengtsson, Patrik Malm, Bo Nordin
Partners: Rajesh Kumar, Centre for Development of Advanced Computing (CDAC), Thiruvananthapuram, Kerala, India; K. Sujathan, Regional Cancer Centre, Thiruvananthapuram, Kerala, India; Andrew Mehnert, Chalmers
Funding: Swedish Governmental Agency for Innovation Systems (VINNOVA); Swedish Research Council; SIDA
Period: 0801-
Abstract: Cervical cancer is a disease that annually kills over a quarter of a million women world-wide. This number could be substantially reduced if women were regularly screened for signs of cancer precursors using the well-established Pap-test. If detected early, these precursors can be treated with a very high rate of success. A problem with the Pap-test is that it requires highly trained cytotechnologists to perform the time consuming visual analysis of the specimen. For over 50 years attempts to automate this process have been made but still no cost effective systems are available.
The CerviScan project is an initiative from the Indian government, managed by the research institute CDAC in cooperation with the Regional Cancer Centre (RCC) in Kerala and CBA in Sweden, aimed at creating a low cost, automated screening system. The system will reduce the number of cytotechnologists needed for population screening by identifying and removing specimen that are clearly normal. A prototype system has been created and used to screen over 1000 specimen. Initial classification results are promising but screening times are still about 10 times longer than what is realistic in a real screening setting. Plans for the next phase of the project, focusing on dedicated hardware, are awaiting the result of a funding application in India.
A sub project on developing improved ways of describing the nuclear chromatin patterns based on new image analysis methods has been spun off and is describe as Project 17. The project has resulted in several recent publications. Patrik Malm defended his PhD thesis closely linked to this project in February 2014.
Jimmy Azar, Anders Hast, Ewert Bengtsson
Funding: TN-faculty, UU
Period: 1001-1410
Abstract: The research was initially part of a VR supported project for grading of prostate cancer. The final part of the research which took place during 2014 extended this to more general ways of describing the architecture of histological tissues.
Immunohistochemistry can facilitate the quantification of biomarkers such as estrogen, progesterone, and the human epidermal growth factor 2 receptors, in addition to Ki-67 proteins that are associated with cell growth and proliferation. We developed a method for the identification of paired antibodies based on correlating probability maps of immunostaining patterns across adjacent tissue sections. Samples from the Human Protein Atlas project were used to test the method.
We also developed a new feature descriptor for characterizing glandular structure and tissue architecture, which form an important component of Gleason and tubule-based Elston grading. The method is based on defining shape-preserving, neighborhood annuli around lumen regions.
These two studies were accepted as journal papers. Jimmy Azar defended his PhD thesis closely linked to this project in October 2014.
Sajith Kecheril Sadanandan, Carolina Wählby
Partners: Johan Elf, David Fange, Alexis Boucharin, Dept. of Cell & Molecular Biology, UU; Klas E. G. Magnusson, Joakim Jaldén, ACCESS Linnaeus Centre, KTH.
Funding: SciLifeLab Uppsala, eSSENCE, VR junior researcher grant to CW
Period: 1210-
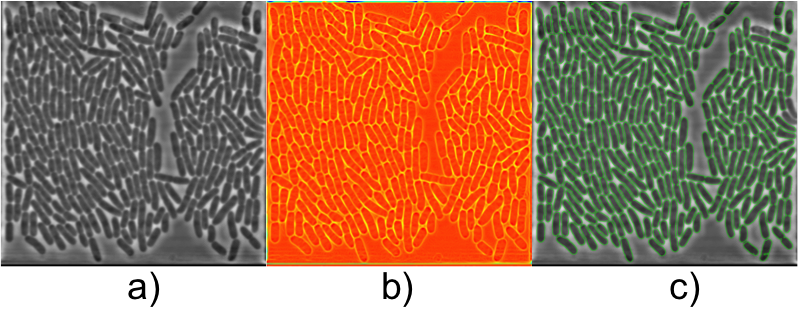
Abstract: Live cell experiments pave way to understand the complex biological functions of living organisms. Most live cell experiments require monitoring of cells under different conditions over several generations. The biological experiments display wide variations even when performed under similar conditions, and therefore need to include large population studied over several generations to provide statistically verifiable conclusions. Time-lapse images of such experiments usually generate large quantities of data, which become extremely difficult for human observers to evaluate. Thus, automated systems are helpful to analysis of such data and provide valuable inference from the experiment. In this work we segment and track E. coli bacteria cells over time. We developed a novel segmentation method, which is fast and robust in delineating bacterial cells in phase contrast microscopy images. The preliminary results (Figure 6) were presented as poster at the International Bioimage Informatics 2014 conference in Leuven, Belgium.
|
Ida-Maria Sintorn, Robin Strand
Partners: Ingela Parmryd, Dept. of Medical Cell Biology, UU; Jeremy Adler, Dept. of Immunology, Genetics and Pathology, UU
Funding: TN-faculty, UU; S-faculty, SLU; VINNMER programme, Swedish Governmental Agency for Innovation Systems
Period: 1101-
Abstract: A cell surface is a highly irregular and rough. The surface topography is however usually ignored in current models of the plasma membrane, which are based on 2D observations of diffusion that really occurs in 3D. In this project, we model diffusion on non-flat surfaces to explain biological processes occurring on the cell surface.
Azadeh Fakhrzadeh, Cris Luengo, Gunilla Borgefors
Partners: Ellinor Spörndly-Nees, Lena Holm, Dept. of Anatomy, Physiology and Biochemistry, SLU
Funding: SLU (KoN)
Period: 1009-
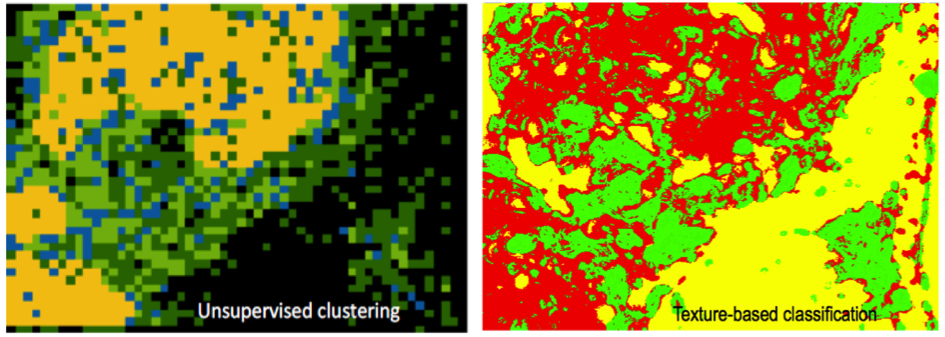
Abstract: Reproductive toxicology is the study of chemicals and their effects on the reproductive system of humans and animals. In reproductive toxicology, there is a strong need to detect structural differences in organs that often have both a complex microscopic structure and function. This problem is further complicated because standard techniques are based on the examination of two-dimensional sections of a three-dimensional structure. The aim of this project is to develop methods to objectively describe microscopic structures of male reproductive organs and to test these in reproductive toxicology research. The project is comparative and includes studies of organs from rooster and mink. We are developing automatic and interactive methods to analyse the relevant structures in the histology images of testis.
Generating sperm in seminiferous tubules is a cyclic process, during which various generations of germ cells in the epithelial layer undergo a series of developmental steps. This cycle is typically subdivided into 12 different stages. We are developing a texture-based classification method to determine each tubule's stage. We are able to distinguish consistently stages if they are grouped into five classes (Figure 7).
Andreas Kårsnäs, Robin Strand, Carolina Wählby, Ewert Bengtsson
Partners: Visiopharm, Hørsholm, Denmark; Clinical Pathology Division, Vejle Hospital, Vejle, Denmark
Funding: NordForsk Private Public Partnership PhD Programme and Visiopharm
Period: 0909-1412
Abstract: The results of analyses of tissue biopsies by pathologists are crucial for breast cancer patients. In particular, the precision of a patient's prognosis, and the ability to predict the consequences of various treatment opportunities before actually exposing the cancer patient, depend on the detection and quantification of biomarkers in tissue sections by microscopy. Experience from the last decade has revealed that manual detection and quantification of biomarkers by microscopy of tissue biopsies is highly dependent on the competencies and stamina of the individual pathologist. The aim of the present PhD project is to develop software-based algorithms that can facilitate the workflow and ensure objective and more precise results of the quantitative microscopy procedures in breast cancer.
Andreas Kårsnäs defended his PhD thesis closely linked to this project in April 2014. A manuscript describing a new method for registering histological images of consecutive sections with different staining by using locally rigid transforms is currently under review and a journal article presenting a histopathological tool for sub-cellular quantification was accepted for publication in the journal Computer methods in Biomechanics and Biomedical Engineering: Imaging & Visualization, in 2014
|
Ingrid Carlbom, Christophe Avenel
Partners: Christer Busch, Dept. of Surgical Sciences, and Anna Tolf, Dept. of Genetics and Pathology, University Hospital
Funding: The Swedish Research Council; Hagstrandska fonden.
Period: 1001-
Abstract: Online Prostate Tissue Grading Tool: Using our OpenSeaDragon-based image selection tool we built an image database of 650 small images from whole mount sections, where each image has one dominant pattern that represents a malignancy grade, precancerous tissue, or benign tissue. With our online grading tool, fourteen internationally prominent pathologists from seven countries are in the process of grading these images according to the Gleason system (Figure 8).
With 50% of the images graded, we see similar grade variations as seen in other studies; for example, in 43% of the cases more than three pathologists disagree. But unlike other studies on intra- and interobserver grading variation, which are based on entire biopsies or whole mounts, each image in our study contains only one dominant pattern, allowing us to identify patterns that cause the discrepancies. Our goal is to establish a consensus for these patterns, thereby promoting international grading standardization.
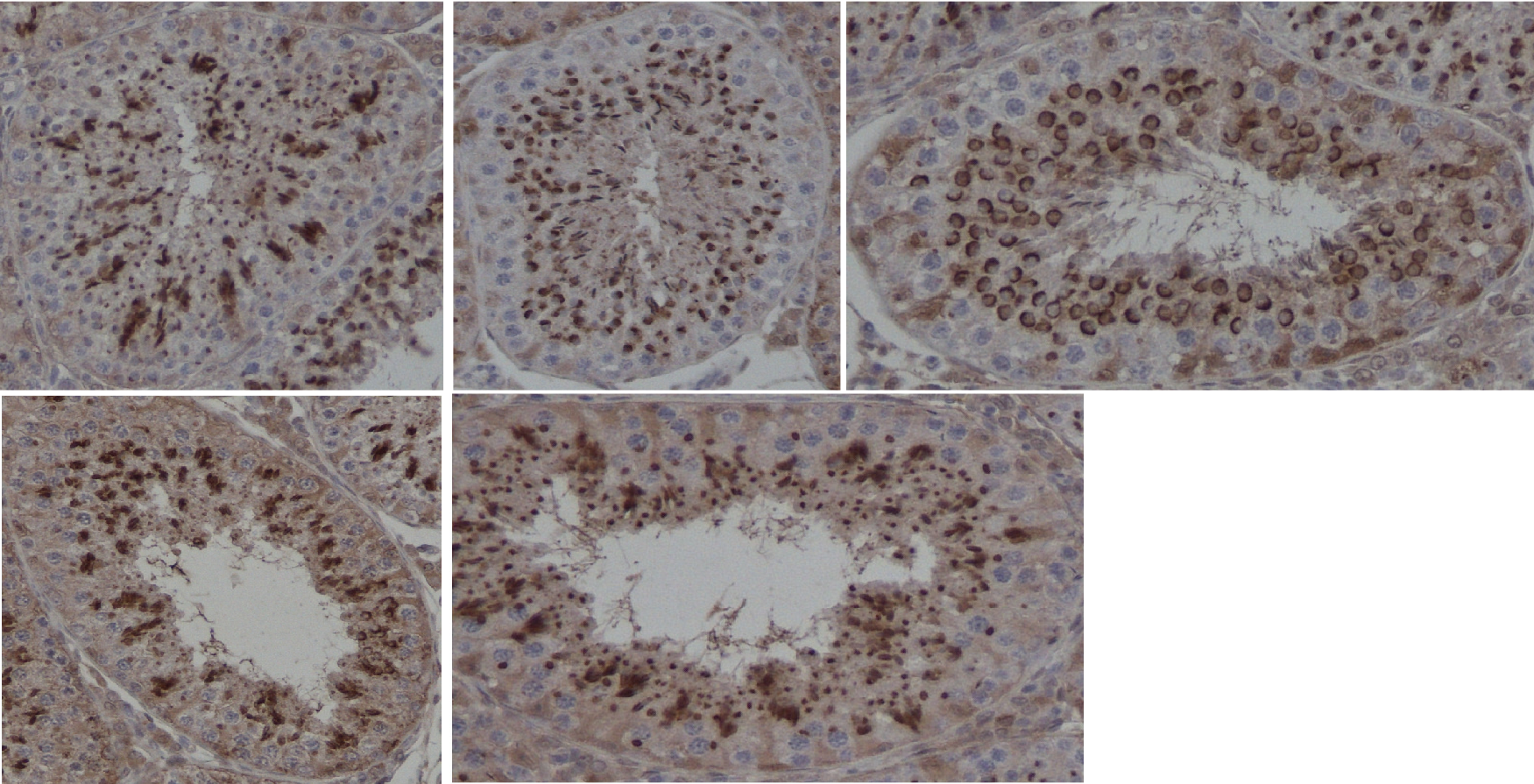
Automatic segmentation of prostate glands and nuclei: Using the stromal density map from the color decomposition of Sirius-hematoxylin-stained tissue, we extract the glandular structures using morphological operations combined with standard image processing operations. With the glandular units as masks, we then extract the epithelium and the lumen from the epithelial density maps. Qualitative analysis of over 5000 glands indicates that the segmentation results in highly accurate representations of the glandular shape, including the epithelium and the lumen (Figure 9a).
Earlier we developed an algorithm to find the epithelial nuclei locations and approximate shapes in the epithelial density maps. We have augmented the algorithm to find a more precise shape of the nuclei using a deformable circular model (Figure 9b).
Consensus-Based Training Dataset: Using the automatically segmented glandular units, we label each gland based on the Gleason Grade produced by one pathologist. This labelling is on a finer scale than the Gleason grades; for example, grade 4 will be separated into fine caliber 4 and cribriform 4. Other grades are also divided to capture differences within the grade. To date we have labelled more than 5000 glands with one pathologist, giving us a dataset for continued development of our automatic grading algorithm. The labels will be updated when we reach a consensus grade with our international panel of experts.
|
Petter Ranefall, Carolina Wählby
Partners: Marcel den Hoed, Manoj Bandaru, Erik Ingelsson, Dept. of Medical Sciences and SciLifeLab, UU
Funding: SciLifeLab Uppsala
Period: 1308-
Abstract: The aim of this project is to identify novel targets for the therapeutic intervention of coronary artery disease. This is done by following-up results from genome-wide association studies in epidemiological studies using a zebrafish model system. Using image analysis we try to identify and characterize causal genes within loci that have so far been identified as associated with coronary heart disease by (high-throughput) screening of atherogenic processes in wildtype and mutant zebrafish, both before and after feeding on a control diet or a diet high in cholesterol. Using confocal microscopy we can image fat accumulation in the zebrafish. Our results confirm that zebrafish larvae represent a promising model system for early-stage atherosclerosis. An abstract has been submitted.
Petter Ranefall, Alexandra Pacureanu, Carolina Wählby, Thu Tran
Partners: Mats Nilsson, Thomas Hauling, Marco Mignardi, Jessica Svedlund, Elin Lundin, Xiaoyan Qian, Dept. of Biochemistry and Biophysics and SciLifeLab, Stockholm University.
Funding: SciLifeLab
Period: 1308-
Abstract: The aim is to create a tool for full resolution image analysis of large images, e.g. slide scanner data, with the possibility of visual examination and interaction at multiple resolutions. The tool is built on a free and open-source framework for visual examination at multiple resolutions with the option to toggle results on or off, such as segmentation masks, classification results, and tissue morphology measurements, using a map view with seamless zooming and panning capabilities, allowing for fast navigation between a full-tissue view and high-resolution sub-cellular observations. The aim is to also have an interface that enables visual/manual selection of regions of interest, target discovery, and understanding of novel spatial relationships within the tissue environment. A bachelor thesis project with the aim of designing the user interface was carried out by the student Thu Tran. This user interface has then been extended with full functionality into a system that we call 'TissueMaps', which is now being evaluated at Mats Nilsson's lab. During 2014 'TissueMaps' has also been presented/demonstrated at some different conferences: BioImage Informatics in Leuven, Belgium, eSSENCE in Umeå, SSBA in Luleå, and the Nordic Symposium on Digital Pathology in Linköping.
Nataša Sladoje, Ewert Bengtsson, Ida-Maria Sintorn
Partners: Joakim Lindblad, Faculty of Technical Sciences, University of Novi Sad, Serbia
Funding: Swedish Governmental Agency for Innovation Systems (VINNOVA); UU TN-faculty
Period: 1308-
Abstract: Within this project we aim at advancing the state-of-the-art methods of digital image processing and analysis, so that some of the societally important and highly challenging and relevant research task in life sciences can be successfully addressed. Our goal is to increase reliability and efficiency, as well as robustness against variations in preparation quality, of computer assisted image analysis in two particular research tracks, related to two applications: (1) Chromatin distribution analysis for cervical cancer diagnostics, and (2) virus detection and recognition in TEM images.
Advances in cyto-patological research combined with the desire for as early as possible cancer detection, have put focus on the analysis of very subtle changes in the chromatin structure of cell nuclei. The application related to recognition and classification of viruses is focused on development of efficient methods adjusted to less expensive equipment, by that increasing applicability and usefulness of the results for a wider community. Efficient utilization of available images data to characterize barely resolved structures, is crucial in both cases. The intention is to use results from resent theoretical work within the framework of discrete mathematics, where knowledge from the field of fuzzy sets is combined with explicit usage of intensity and shape information in images. This theoretical framework provides methods which enable preservation and efficient usage of information, aggregate information of different types, improve robustness of the developed methods and increase precision of the analysis results. Based on conducted studies, the developed framework has shown high potential in applications where efficient utilization of information is of importance and where reliability of the analysis results and their timely delivery are both required.