Xavier Tizon, Roger Lundqvist, Gunilla Borgefors, Ewert Bengtsson
Funding: Swedish Foundation for Strategic Research, VISIT programme
Period: 9707-0208
Partners: Lennart Thurfjell, Personal Chemistry AB, Uppsala; Örjan Smedby, Dept. of Medicine and Care, Linköping University; UU Hospital; Dept. of Nuclear Medicine and Dept. of Neuroradiology, Karolinska Hospital, Stockholm; Dept. of Nuclear Medicine, The Prince of Wales Hospital, Sydney, Australia
Abstract: This is a long-term project where the overall objective is to develop methods to support diagnosis based on 3D medical images. The data sets provided by the latest imaging techniques can sometimes be confusing to interpret for the physicians, mainly because of the complexity of the 3D structures visualised and the lack of sufficiently informative visualisation techniques. The images need to be presented to the physicians in such a way that they can be easily and correctly interpreted, but without removing essential information. In particular, we concentrate on the development of new methods for registration, diagnosis, visualisation, and segmentation.
Fusion of brain images
One part of the project was focused on analysis of PET, SPECT, MR, and CT
brain images. We developed voxel-based registration methods, both for rigid
registration of data from different examinations of the same patient and
for non-rigid registration of data from different individuals.
The rigid registration methods can be used to fuse information acquired
from different imaging modalities to improve medical diagnosis. One example
is from patients suffering from epilepsy where an examination may contain
both SPECT and MR scans and a fusion of the two scans enables accurate
determination of the location of the epileptic foci.
Another important part throughout the fusion project has been a
computerised brain atlas, which maps the human brain into anatomical and
functional regions. Improved methods for atlas-based analysis and more
accurate image registration methods have been developed. The brain atlas
technique was used in a project aiming to quantify the effects from carotid
endarterectomy on patients suffering from carotid artery stenosis. Selected
vascular brain atlas regions were used to measure the differences between
preoperative and postoperative SPECT scans. Finally, the measured
differences were correlated to other observed variables describing the
outcome of the operation. Previously, the brain atlas was used for
discrimination from SPECT scans between patients suffering from Alzheimer's
disease and normal controls subjects.
Another development result has been improved methods for combined
3D visualisation of volume images. The new methods enables visualisation of
information from different imaging modalities together in the same rendered
image. Furthermore, methods has been developed to incorporate atlas
structures into the 3D visualisation, which enables more accurate
localisation of different features in the image data.
In November 2001, this work was presented and defended as a PhD thesis by
Lundqvist. During 2002, he has worked on making the implementation of the
methods in the atlas more robust and user friendly, on clinical
verification of the methods and on finalising some publications documenting
the results.
Arteries-veins separation in magnetic resonance angiography images
Another part of the project aimed at selecting a subset of volumetric data,
and to present it in such a way as to make diagnosis easier. As an example,
in magnetic resonance angiography (MRA), it is of great interest to be able
to separate arteries from veins. This problem is not trivial, because the
vessels can lie in close parallel throughout the image. Our algorithm
extends the concept of binary connectedness by using a grey-level
connectedness approach using fuzzy sets. As a start set we used small sets
of voxels marked by the user. Good user interaction possibilities,
portability and reusability were important concerns in this project. That
is why we chose to use public domain packages: the Visualization Toolkit
(VTK) and the Insight Segmentation Toolkit. The work was published in
``Journal of Magnetic Resonance Imaging'' during this year.
Xavier Tizon, Gunilla Borgefors
Funding: Swedish Foundation for Strategic Research, VISIT programme
Period: 0209-
Partners: Lars Johansson, Håkan Ahlström, Dept. of Oncology, Radiology, and Clinical Immunology, UU Hospital
Abstract: As part of a large clinical trial launched in order to study the causes of arteriosclerosis, a large number of volunteers will take part in a whole-body MR angiography study, using a protocol developed at the UU Hospital. The goal of the project is to develop Image Analysis tools to derive global measures of arteriosclerosis, from the characteristics of a limited portion of the arterial tree. The project will be split in four sub-tasks:
- 1.
- Registration of the sub-volumes. The acquisition technique produces four MRA volumes that correspond to the head and upper torso, the abdomen, the upper legs and the lower legs. In order to be able to apply the subsequent image analysis procedure, they have to be assembled into one single image. The challenge here is the strong non-linearity of the imaging process, which shows considerable geometric distortion near the borders of the volume. This has to be corrected automatically, for example using a phantom approach, see Figure 2, left.
- 2.
- Segmentation and identification of the arterial structure. Once the full volume has been reconstructed, the next step is the segmentation of the arteries from the rest of the visible structures. See Figure 2, right, for an unsegmented MRA image. Some methods developed in other projects, e.g., Project 1 will be used. The segmented arteries must then be identified, and only the interesting parts of the artery tree should be kept.
- 3.
- Geometric measures to give an estimate of plaque burden. The geometry of the tubular structure of the arterial tree has to be studied. Irregularities in the diameter along the central line of the vessel may be a candidate measure of the plaque burden, but other indicators may also be interesting.
- 4.
- Clinical testing. It is important, finally, to evaluate the clinical significance of the potential measures derived in Step 3. A clinical study will help to choose which one is the best for the physician to use. This part involves the writing of user-friendly and robust software, that would have to be used in different environments

|
Ewert Bengtsson, Roger Hult
Funding: VINNOVA, UU TN-faculty
Period: 9501-
Partners: Depts. of Neuroradiology and Clinical Neurophysiology, Karolinska Institute and Hospital, Stockholm; Dept. of Physics, Stockholm University; PET Centre, UU; Lennart Thurfjell, Personal Chemistry AB, Uppsala
Abstract: The objective of this project is to develop new tools for analysis and visualisation of neuroimaging data. These tools are partly integrated in a computerised brain atlas. The work during the last years has been focused on segmentation of MR images and visualisation of functional information on the segmented volumes. A segmentation method based on connectivity analysis and morphology has been developed. The goal is to have a robust three-dimensional method for segmentation of the brain in MRI data. The project is carried out in close collaboration with the medical partners.
Ewert Bengtsson, Roger Hult
Period: 0105-
Partners: Håkan Hall, Ingrid Agartz, Dept. of Clinical Neuroscience, Karolinska Institute and Hospital, Stockholm; Stefan Arnborg, NADA, KTH, Stockholm
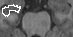
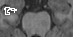
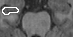
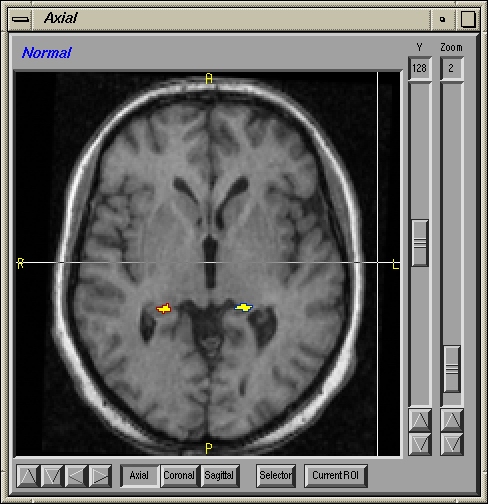
Abstract: This is part of a larger project with the objective to perform research on the brain to find new solutions and treatments for brain diseases. The project HUBIN (Human Brain Informatics) is an interdisciplinary project that for the time being is focusing on schizophrenia and started in 1998. Within HUBIN there are several projects and the aim for the image analysis project is to be supportive in using the software BRAINS2. Tools for segmenting structures in the brain are also being developed and implemented in the BRAINS2 software. The software uses ANN (artificial neural networks) to segment structures in the brain. A tool for segmenting the hippocampus is under development. See Figures 3 and 4. The project is carried out in close collaboration with the medical partners.
Figure 3: Left: A manually segmented slice of the right hippocamopus. Middle: A segmented slice of the hippocampus using the original automatic segmentation method. Right: A segmented slice of the hippocampus using the improved automatic segmentation method.
Figure 4: Left: A slice showing the two hippocampi segmented. Right: A 3D view of the two hippocampi.
Pascha Razifar, Ewert Bengtsson
Period: 0110-
Funding: PET Centre, UU; UU TN-Faculty
Partners: Mats Bergström, Harald Schneider, PET Centre, UU
Abstract: Within this project existing methods for visualising 3D anatomical information and methods for visualising multidimensional features as scatter plots etc. will be extended and integrated into a coherent tool for interactive exploitation of these highly complex data. One concept that will be investigated is how the anatomical information can be visualised in the clusters pace through spectral and temporal methods. The work will be based on sequences of PET images from various tracer studies.
The motivation for this work is the highly complex data sets that are obtained from PET studies and which are difficult to fully interpret with currently available tools. New ways of interacting with the data that preserves the anatomical context at the same time as it allows flexible exploitation of the many feature dimensions should have a potential of providing most useful tools. The project was defined during the second half of 2001 and is thus in an early phase.
One-way of understanding the nature of PET images and thus how they can be effectively interpreted is to create synthetic PET images with full control over the correlated noise processes, etc. During 2002, a program that can generate synthetic PET images has been developed. One result of this work is the application of the auto-correlation function (ACF) for verification of the precision of the reconstruction algorithm in the actually used PET cameras.
Xavier Tizon, Gunilla Borgefors
Funding: Swedish Foundation for Strategic Research, VISIT programme
Period: 0108-
Partners: Sven Nilsson, Dept. of Oncology, Radiology, and Clinical Immunology, UU Hospital
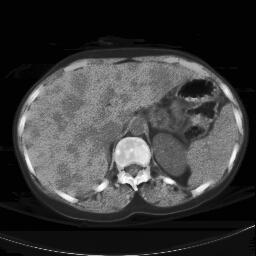
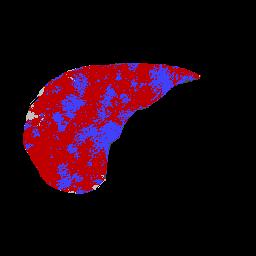
Abstract: The liver is a common site of metastatic disease. The diagnosis of liver metastases in patients at high risk is straightforward, but estimating the gravity is more problematic. We investigate the use of CT scans, see Figure 5, left. The first problem is to segmenting the liver itself in the CT scan, the second is segmenting the metastases in the liver to evaluate their relative volume. In this pre-study, we investigated the possible methods available to us, see Figure 5, right. The algorithms have to be 3D in nature, but are allowed to include some basic user interaction. Further work implies the use of level-set methods, and/or fuzzy topological algorithms.
Figure 5: Left: Liver metastasis as seen on a CT scan slice of a patient. Right: A primary segmentation result using the mean-shift algorithm.
Patrick Karlsson, Ewert Bengtsson
Funding: UU TN-faculty
Period: 0212-
Partners: Christer Busch, Dept. of Genetics and Pathology, The Rudbeck Laboratory, UU; Kenneth Wester, Depts. of Surgical Sciences, Experimental Urology and Genetics and Pathology and Biomedical Radiation Sciences, The Rudbeck Laboratory, UU
Abstract: Quantification of microvessel density in prostatic carcinoma is often done by calculating the number of and/or the area outlined by the endothelial cells. The result is expressed in relation to the quantified tumour area. Automated image analysis quantification of microvessel density has proven to correlate well with manual quantification done by a pathologist. We have observed that the microvessel-pattern is highly dependent on the tissue architecture, which in turn depends on the distribution and size of the pros tate glands. This indicates that, besides microvessel density, also microvessel pattern may be of prognostic importance in prostate cancer diagnostics. In this project, a new approach to microvessel quantification is tested and evaluated in comparison to malignancy grading and classical microvessel density.
Carolina Wählby, Ewert Bengtsson
Funding: Swedish Foundation for Strategic Research, VISIT program
Period: 9806-
Partners: F. Erlandsson and A. Zetterberg, Dept. of Oncology/Pathology, CCK, KI, Stockholm
Abstract: Shape and distribution of various subcellular structures and components can be observed by immunostaining and insitu-hybridisation of fluorescent markers followed by fluorescence microscopy in three dimensions. The 3D images are acquired by making non-invasive serial optical sections of the object. Studies of the distribution of signal factors involved in the cell cycle control indicate that minor changes in the signalling systems are the first signs of cancer transformation and tumour formation. Understanding the 3D organisation of normal and transformed cell-nuclei is therefore of great interest as a new approach to understanding the pathways of cancer. A new technique for sequential immunofluorescence staining has been developed in cooperation with the Dept. of Oncology/Pathology, Div. of Tumour Cytology, CCK, Karolinska Hospital and Institute. New methods for analysis of these multi-dimensional data are currently tested. The methods include image registration, semi-automatic segmentation, and normalisation. The aim of the project is to examine the relationships between changes in the signalling systems within individual cells, either in culture or in sections of tumours. A second part of this project is carried out in Gustavsson's research group at the Dept. of Signals and Systems, Chalmers University of Technology, Göteborg.
Carolina Wählby, Joakim Lindblad, Mikael Vondrus, Ewert Bengtsson
Funding: Amersham Biosciences, Uppsala, Cardiff, UK; UU TN-faculty; SSF through the VISIT programme
Period: 9902-
Partners: Lennart Björkesten, Amersham Biosciences, Uppsala; Stuart Swinburne, Simon Port, Alla Zaltsman, Gareth Bray, and Dietrich Ruehlmann, Amersham Biosciences, Cardiff, UK
Abstract: The interaction with and effect of potential drugs on living cells can be observed by fluorescence microscopy. High throughput methods for analysis of cells can be used as a tool in the drug discovery process. The overall objective of this project is to develop image analysis methods for segmentation, feature extraction and classification of cells and subcellular structures in fluorescence microscopy images.
Segmentation of cytoplasms
The cell nucleus has a well-defined shape and is relatively easy to detect.
The cytoplasm is however more complex. The first goal of this project was
to develop a fully automatic method for cytoplasm segmentation. The present
algorithm, inspired by literature and previous experience, consists of an
image pre-processing step, a general segmentation and merging step followed
by a quality measure and a splitting step. By training the algorithm on one
image, it is made fully automatic for subsequent images created under
similar conditions. This method was presented at an internal Amersham
Pharmacia Biotech R&D conference in Uppsala in late 1999. During 2000 the
algorithms were improved through a more elaborated shape analysis and a
more consistent feature extraction and quality evaluation step. The results
were presented at an international conference in 2001 and documented in a
journal paper that was published in 2002.
Classification of Rac1 activation
Based on the experience from the segmentation of cytoplasms, a more problem
specific project was initiated in cooperation with Amersham Biosciences in
Cardiff, UK, in 2002. The aim of this project was to develop algorithms
enabling fully automatic, real-time segmentation and analysis of
fluorescence images of cells so as to quantitatively estimate the IGF-1
induced translocation of GFP-Rac1 to the cellular membrane for individual
cells. See Figure 6. Due to the ultimate goal of analysing
one image containing roughly 200 cells in less than two seconds, effort was
taken not to use algorithms of high time complexity. The results were
documented in a journal paper to be submitted for publication.
Figure 6: CHO-hIR cells expressing GFP-Rac1 fusion protein, imaged on IN Cell Analyzer, Amersham Biosciences, Cardiff, UK. Cytoplasms, before (left) and 4.3 min after (right) incubation with IGF-1. The translocation of GFP-Rac1 appears as bright formations along the edges of the cells. This translocation can be quantified by image analysis.
Ida-Maria Sintorn, Gunilla Borgefors
Funding: SLU S-faculty
Period: 0111-
Partners: Mohammed Homan, Cecilia Söderberg-Naucler, Centre for Molecular Medicine, Karolinska Hospital, Stockholm
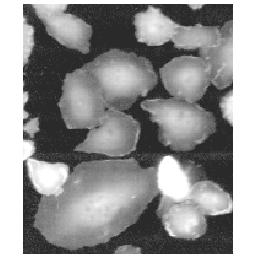
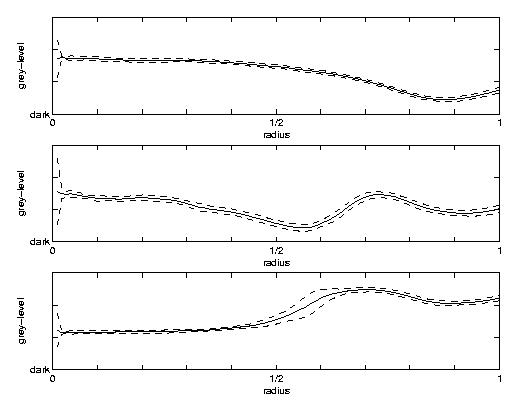
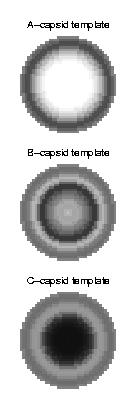
Abstract: Human Cytomegalo Virus (HCMV) is a rather unexplored virus belonging to the herpes virus family. The goal of this project is to segment, classify, and describe virus capsids at three different maturation stages from transmission electron microscopy (TEM) images of an infected cell nuclei. The virus capsids are to be classified as immature, in between, or mature. The segmentation is done by template matching for one class at a time. The templates, see Figure 7, right, were produced from the images of a number of typical particles of each class. Each class is then described by producing a normalised mean radial grey-tone profile, see Figure 7, left, from the capsids segmented to each class, i.e., the mean grey-level for all pixels at the same distance from the particle edge was computed for all distances.
Figure 7: Left: Class grey-level profiles and standard deviation for the three classes of capsid maturation. The radius is ranging from 0, being the centre, to 1 being the edge of the capsid shelf. The solid lines correspond to the grey-levels at each radius and the dashed lines to the grey-level +/- 2 standard deviations. Right: Templates of radius 20 constructed from the class grey-level profiles.
Ingela Nyström, Ida-Maria Sintorn
Funding: UU TN-faculty, SLU S-faculty
Period: 9906-
Partners: Stefan Höglund, HIV structure group, Dept. of Biochemistry, UU
Abstract: Sample specimens of HIV-1, and HIV-1 treated with the tripeptide GPG, are studied in series of different tilt angles from +60° to -60° with a goniometer in transmission electron microscopy (TEM). A tilt series consists of 25-40 electron micrograph projections. Each of these is digitised. Thereafter, the digital images are aligned, using the coordinates of (some of) the added gold particles (10 nm) as reference points. The 3D reconstruction is made as a series of 2D reconstructions, each a combination of the radius-weighted Fourier transform of one pixel line of each micrograph. The 3D reconstructions are typically of size 256 × 256 × 80 voxels (appr. 5 Mbyte). The goal of the project is to achieve 3D reconstructions of intact and GPG treated single HIV-1 particles for comparison and quantitative analysis through, e.g., convex hull computat ions. The effects of GPG treatment of HIV-1 is presented in the November 2002 issue of Antimicrobial Agents and Chemotherapy.