
|
Identification of Highly Pathogenic Viruses in Transmission Electron Microscopy Images Gustaf Kylberg, Ida-Maria Sintorn, Ewert Bengtsson, Gunilla Borgefors
Partner: Vironova AB; Delong Instruments, Brno, Czech Republic; Ali Mirazimi, Kjell-Olof Höglund, Centre for Microbiological Preparedness; Swedish Institute for Infectious Disease Control (SMI)
Funding: Swedish Civil Contingencies Agency (MSB); Swedish Defense Materiel Administration (FMV); Swedish Agency for Innovative Systems (VINNOVA). Eurostar project E!6143
Period: 0801-
Abstract: This project aims at automating the virus identification process in high resolution TEM images. This, in combination with Project 6 create a rapid, objective, and user independent virus diagnostic system. The identification task consists of method development for segmenting virus particles with different shapes and sizes and extracting descriptive features of both shape and texture to enable the classification into virus species. Texture features such as variants of Local Binary Patterns and Regional Moments (filter banks constructed from orthogonal moments), are being evaluated on virus textures as well as other texture datasets to get a deeper understanding of the discriminant power of the features under different conditions. A paper evaluating the discriminating power and noise robustness for Local Binary Pattern variants was published during 2013, and a poster about the project was presented at the Microscopy Conference in Regensburg, Germany in August.
The miniTEM Project - Development of a Desk-top TEM with Automated Image Acquisition
Gustaf Kylberg, Ida-Maria Sintorn, Ewert Bengtsson, Gunilla Borgefors
Partner: Vironova AB; Delong Instruments, Brno, Czech Republic
Funding: Eurostar project E!6143
Period: 1107-
Abstract: Transmission electron microscopy (TEM) is an important clinical diagnostic and material analysis tool. Transmission electron microscopes are expensive, complex, sensitive and bulky machines, often housed in specially built rooms to avoid vibrations affecting the imaging process. They are to a very large extent manually operated, meaning that an expert in electron microscopy and preferably also in the application at hand needs to perform the analysis at the microscope, an often very time consuming task.
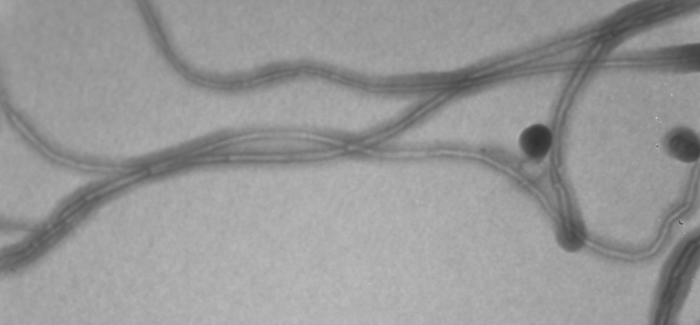
This project aims at developing the miniTEM, shown in Figure 3(left), a desk-top low voltage TEM designed for imaging biological samples, with a high degree of automation regarding instrument alignment, image acquisition and analysis. The goal is a small, cheap, robust, and easy to use system that requires no more training than any simple lab equipment, and can be hosted in any office or lab (even mobile).
Automating the image acquisition process is key for reducing the manual input and making the imaging and analysis more objective. A few different options for automated image acquisition are being developed and will be incorporated in the instrument. The first is acquisition of images at random positions on the grid. The second is to search for a specific structure/object and only acquire (store) the images containing the structure/object of interest. The third is similar to the second approach but embedded in a multi-scale approach with the goal to make the acquisition more efficient.
The very first images from the miniTEM were acquired at the end of 2013. An example image of nanotubes with an approximate thickness of 15nm are shown in Figure 3(right). Work on optimizing the sample preparation procedure for improved electron transmittance was presented at the Microscopy Conference in Regensburg, Germany in August.


|
Detection and Localization of Florescent Signals in STORM Data Using Compressed Sensing
Omer Ishaq, Alexandra Pacureanu, Carolina Wählby
Partners: Johan Elf, Gustaf Ullman, Fredrik Persson, Dept. of Cell & Molecular Biology, UU
Funding: SciLifeLab Uppsala, eSSENCE, VR junior researcher grant to CW
Period: 1211-
Abstract: Stochastic optical reconstruction microscopy (STORM) is a super-resolution microscopy image acquisition technique for single-molecule localization. Like other stochastic super-resolution microscopy techniques it incorporates a trade-off between spatial- and temporal-resolution. Recently, a compressed-sensing (CS) based variant of STORM, called FasterSTORM, has been developed which substantially increases the temporal sampling of a stack of STORM image frames. This improvement is realized by increasing the density of activated fluorophores in each frame, followed by a subsequent CS-based retrieval of single-molecule positions even with overlapping fluorescent signals. However, the CS-based retrieval/decoding step is time consuming and can take as much as three hours for each image frame. We have accelerated the FasterSTORM method through parallel processing on multi-core processors. Additionally, we have tested and tried a number of L1-solvers for CS-based recovery of molecule positions. A paper comparing convex and greedy solvers and evaluating the sensitivity of the FasterSTORM to estimation bias of the point spread function (PSF) was submitted to a conference. We are in the process of comparing the performance of the Faster STORM against a wavelet-based approach to localize fluorescent signals in time-lapse images of bacterial cells.
In Situ Sequencing of mRNA
Carolina Wählby, Alexandra Pacureanu, Petter Ranefall
Partners: Mats Nilsson, Rongqin Ke, Marco Mignardi, Thomas Hauling, SciLifeLab Stockholm
Funding: SciLifeLab Uppsala; TN-faculty, UU
Period: 1109-
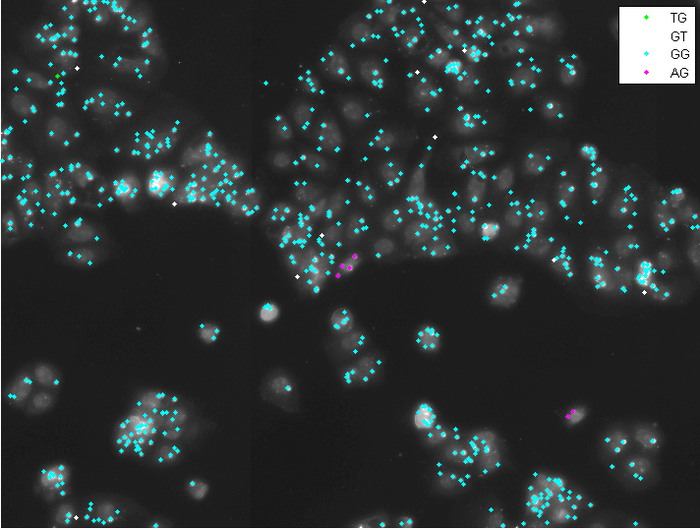
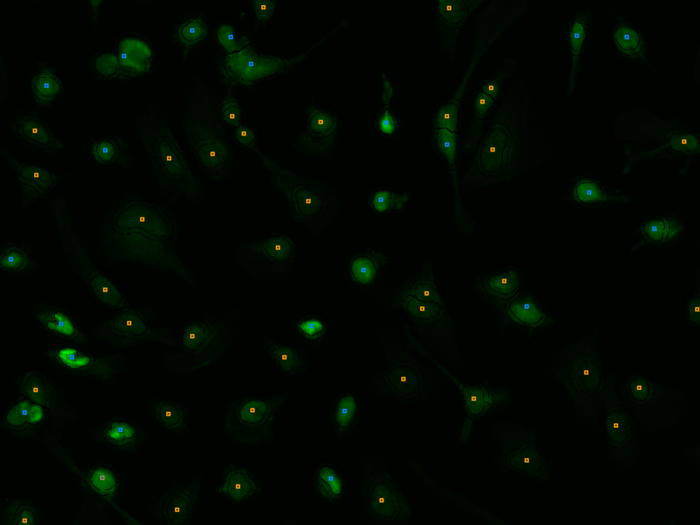
Abstract: Profiling of gene expression is prerequisite for understanding the function of cells, organs and organisms, in health and disease. The sequencing techniques currently in use rely on homogenization of the samples. Therefore, the obtained information represents either the average expression profile of the tissue sample or expression profiles of isolated single cells. Our collaborators have developed a new molecular method, enabling in situ sequencing of mRNA, so that protein expression can be observed directly in cultured cells or tissue samples. We have developed image analysis tools for automated analysis of sequencing data, mapping, and visualization of gene expression patterns (Fig. 4). In 2013 we published a paper in Nature Methods and a conference paper focusing on the image analysis was accepted for publication in proceedings of the IEEE International Symposium on Biomedical Imaging (ISBI), Beijing 2014.
|
Evaluation of the Effect of Compaction Oligonucleotides on the Strength and Integrity of Florescent Signals
Omer Ishaq, Petter Ranefall, Carolina Wählby
Partners: Carl-Magnus Clausson, Linda Andersson, Ola Söderberg, Dept. of Immunology, Genetics and Pathology
Funding: SciLife Lab Uppsala
Period: 1310-
Abstract: Rolling circle amplification (RCA) performs nucleic acid replication for rapid synthesis of multiple concatenated copies of circular DNA. These molecules can be visually observed through the use of florescent markers. Moreover, the introduction of a compaction oligonucleotide during RCA results in brighter and more compact signals. The project aims to evaluate the effect of compaction oligonucleotides on the strength and integrity of florescent signals.
Skeleton-Based Vascular Segmentation at Interactive Speed
Kristína Lidayová, Hans Frimmel, Ewert Bengtsson
Partner: Örjan Smedby, Chunliang Wang, Center for Medical Image Science and Visualization (CMIV), Linköping University
Funding: VR grant to Örjan Smedby
Period: 1207-
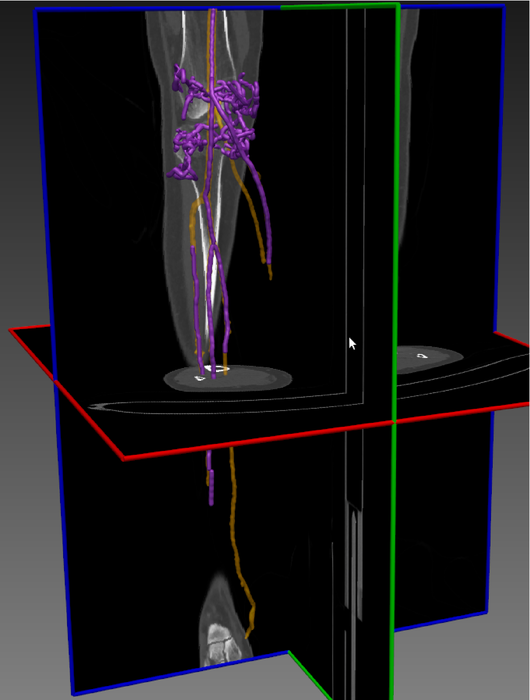
Abstract: Precise segmentation of vascular structures is crucial for studying the effect of stenoses on arterial blood flow. The goal of this project is to develop and evaluate vascular segmentation, which will be fast enough to permit interactive clinical use. The first part is the extraction of the centerline tree (skeleton) from the gray-scale CT image. Later this skeleton is used as a seed region (Figure 5). The method should offer sub-voxel accuracy.
During the last year we improved the software for fast vessel centerline tree extraction. The method has been tested on several CT datasets and the results look promissing. Generally all main vessel centerlines are detected, but some improvement needs to be done in order to remove some false positive centerlines.
|
Computational Methods for Quantification in Neural Stem Cells
Alexandra Pacureanu, Carolina Wählby, Martin Simonsson
Partners: Karin Forsberg-Nilsson, Tanja Paavilainen, Soumi Kundu, Grzegorz Wicher, Lisa Rebello, Anqi Xiong, Tobias Bergström, Dept. of Immunology, Genetics and Pathology, Rudbeck Laboratory, SciLifeLab Uppsala
Funding: SciLifeLab Uppsala
Period: 1210-
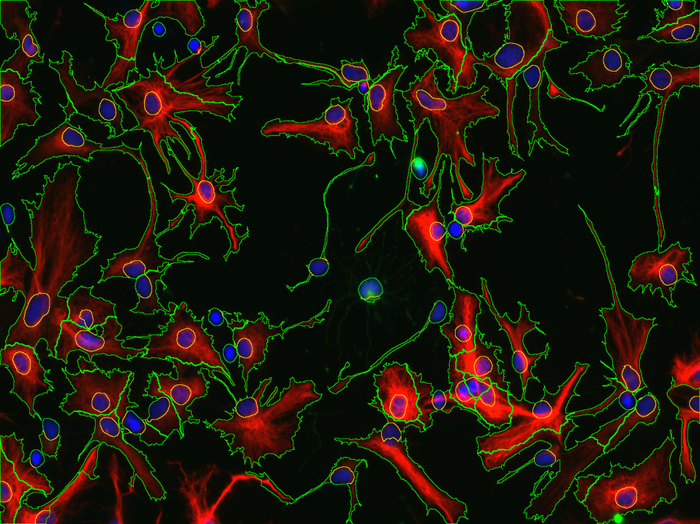
Abstract: Neural stem cells are the building blocks of the nervous system. In the view of finding better treatments for neurodegenerative diseases and for deeper understanding of mammalian development, our collaborators are investigating how neural stem cells proliferate and differentiate and which factors govern these processes. For these studies, thousands of images of cell cultures need to be quantitatively analyzed, in order to determine for example how effective are various techniques for control of the stem cells differentiation. Based on CellProfiler and CellProfiler Analyst, we have developed methods for automatic analysis of these images (Fig. 6). In 2013, the master thesis of Tanja Paavilainen has been successfully completed and we continued the collaboration with researchers from the Karin Forsberg group. For example, we have been working together with Tobias Bergström on quantification of the OLIG2 expression in different glioma cell lines and with Soumi Kundu on blood vessels segmentation.
|
SciLifeLab Cancer Stem Cell Program
Carolina Wählby, Ida-Maria Sintorn
Partners: Sven Nelander, Karin Forsberg-Nilsson, Irina Alafuzoff, Ulf Landegren, Anna Segerman, Tobias Sjöblom, Lene Urborn and Bengt Westermark, Department of Immunology, Genetics and Pathology and SciLifeLab, UU, Bo Lundgres, the Karolinska Institute and SciLifeLab, Stockholm, Rebecka Jörnsten, Chalmers, Gothenburg, and Göran Hesselager, UU Hospital, Uppsala
Funding: AstraZeneca-Science for Life Laboratory Joint Research Program
Period: 1303-
Abstract: The SciLifeLab Cancer Stem Cell Program is a cross-platform initiative to characterize cancer stem cells (CSCs). Previously, the development of drugs targeting the CSC population in solid tumors has been curbed by the lack of valid cell model systems, and the complex genetic heterogeneity across tumors, factors that make it hard to assess new targets or predict drug responses in the individual patient. To solve these problems, our aim is to develop a biobank of highly characterized CSC cultures as a valid model of cancer heterogeneity. We will combine mathematical and experimental approaches, including image-based high-throughput cell screening, to define the spectrum of therapeutically relevant regulatory differences between patients. This will help elucidate mechanisms of action and enable accurate targeting of disease subgroups. During 2013, patient data was collected, and a number of primary cell lines were established. Cultured cells were exposed to a different treatments and doses (more than 2500 different treatments per cell line), and imaged by fluorescence as well as bright-field microscopy, and current focus is on extracting meaningful morphological descriptors from the image data.
Endothelial Cell Segmentation of the Cornea of Human Eyes
Bettina Selig, Cris Luengo
Partners: Bernd Rieger, Quantitative Imaging Group, Delft University
of Technology, Netherlands; Koen Vermeer, The Rotterdam Eye Hospital, The Netherlands
Funding: S-faculty, SLU
Period: 1103-
Abstract: The corneal endothelium plays a key role in maintaining the
transparency of the cornea. Because the cells in the endothelium do not
regenerate, the cell density decreases with age; this reduces its ability
to maintain the processes needed to keep the cornea transparent. Thus, being
able to measure this density in patients is very important. The endothelium
can be imaged by specular microscopy or by confocal scanners, and measurements
can be obtained manually, automatically with manual corrections, or fully
automatically with current software (e.g., Nidek's NAVIS). Unfortunately, the
results of the automatic methods are often useless, especially at low cell
densities. Together with the Rotterdam Eye Hospital, we have developed a fully
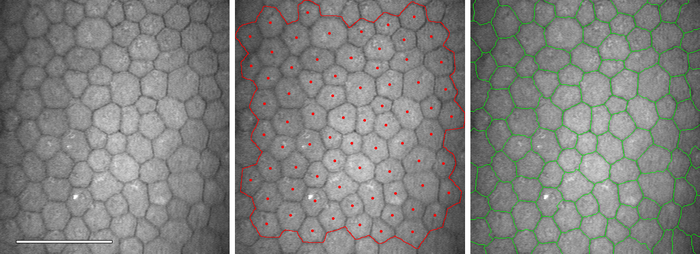
automatic method to segment individual cells in the corneal endothelium. The
result of the method (see Figure 7) can be used to determine
the cell density, but also other parameters of interest, like pleomorphism
(cell shape) and polymegathism (cell size variation). Our segmentation method produces a segmentation that matches a manual segmentation reasonably well, for a wide range of cell densities and image qualities. These results will be
published during 2014.
|
CerviScan
Ewert Bengtsson, Patrik Malm, Hyun-Ju Choi, Bo Nordin, Andrew Mehnert
Partners: Rajesh Kumar, Centre for Development of Advanced Computing (CDAC), Thiruvananthapuram, Kerala, India; K. Sujathan, Regional Cancer Centre, Thiruvananthapuram, Kerala, India
Funding: Swedish Governmental Agency for Innovation Systems (VINNOVA); Swedish Research Council; SIDA
Period: 0801-
Abstract: Cervical cancer is a disease that annually kills over a quarter of a million women world-wide. This number could be substantially reduced if women were regularly screened for signs of cancer precursors using the well established Pap-test. If detected early, these precursors can be treated with a very high rate of success. A problem with the Pap-test is that it requires highly trained cytotechnologists to perform thetime consuming visual analysis of the specimen. For over 50 years attempts to automate this process have been made but still no cost effective systems are available.
The CerviScan project is an initiative from the Indian government, managed by the research institute CDAC in cooperation with the Regional Cancer Centre (RCC) in Kerala and CBA in Sweden, aimed at creating a low cost, automated screening system. The system will reduce the number of cytotechnologists needed for population screening by identifying and removing specimen that are clearly normal. A prototype system has been created and used to screen over 1000 specimen (Fig. 8). Initial classification results are promising but screening times are still about 10 times longer than what is realistic in a real screening setting. Plans for the next phase of the project, focusing on dedicated hardware, are under way and are currently awaiting the result of a funding application. In Sweden, Ewert Bengtsson and Patrik Malm at CBA have in collaboration with Andrew Mehnert and students at MedTech West, Chalmers, been working on developing improved texture measures that are based on pseudo-3D information generated by imaging cells as focus stacks. This work is ongoing but has already led to two conference publications with a third manuscript on the way. Other work at CBA includes methods for nucleus segmentation, debris removal and field of view grading. Also, advanced procedural methods for synthetic Pap-smear image generation have been developed and published. Currently, a study aimed at determining the optimal optical resolution for a future system is taking place. Preliminary results of this study have been composed into a manuscript and submitted for conference publication.
The work has been summarized in a thesis dubbed "Image Analysis in Support of Computer-Assisted Cervical Cancer Screening" . The thesis was defended February 7, 2014 by Patrik Malm.
|
Automated Tissue Image Analysis using Pattern Recognition
Ewert Bengtsson, Anders Hast, Jimmy Azar
Funding: TN-faculty, UU
Period: 1001-
Abstract: The research was initially part of a VR supported project for grading
of prostate cancer; the final part of the research will focus on
analysis of samples from the Human Protein Atlas project aiming at
developing methods that can be used for computer assisted image
analysis of the huge database of images generated in that project.
In particular, image analysis methods will be developed for
histopathology, initially with applications to grading of prostate
cancer, but with extension to structural analysis of breast tissue
samples. The methods will be verified using paired antibody
evaluations.
Detection and Classification of Malaria Infected Cells by LED Spectral Microscopy
Carolina Wählby
Partners: Jeremie Zoueu, Olivier Bagui, Dept. Genie Electrique et Electronique, Institut National Polytechnique, Felix Houbhouet-Boigny, Cote d' Ivoire
Period: 1109-
Abstract: This project aims to propose an effective optical device based on LED spectral microscopy, which will be low cost, fast and easy to use in the diagnosis of human malaria parasites, especially because the sample will not need any special preparation or staining and the data will be automatically processed to provide real-time diagnosis of the type of the parasite, the parasitic density and its age for an effective prescription. The collaborative project was initiated by a 3-month visit by Olivier Bagui, where we focused on the development of efficient segmentation methods for unstained images of blood cells. An efficient segmentation approach was developed using CellProfiler. Segmentation masks could thereafter be used to extract per-cell measurements for further exploration of spectral information on single cells.
Studying Exocytosis by Time Lapse Microscopy
Martin Simonsson, Carolina Wählby
Partners: Anne Wuttke, Dept. of Medical Cell Biology, UU
Funding: SciLifeLab Uppsala, eSSENCE, VR junior research grant to CW
Period: 1211-
Abstract: Insulin secreting cells perform exocytosis and this can be detected with a GFP-modified protein as an increase in fluorescence signal. Time-lapse sequences are acquired with a time interval of one second during one hour, observing changes in fluorescence signaling at different treatments of the cells. This results in huge data sets with more than 3000 images for a single experiment. The focus of this project is to extract relevant information from the image data and in an efficient way analyze and visualize the data. Preliminary results were presented in a PhD thesis by our collaborator Anne Wuttke, and a manuscript is in preparation.
Tracking of Unstained Cells in Microfluidic Systems
Sajith Kecheril Sadanandan, Martin Simonsson, Carolina Wählby
Partners: Johen Kreuger, Sara Thorslund, Gradientech AB, Uppsala
Funding: SciLifeLab Uppsala; eSSENCE; Dept. of IT, UU
Period: 1108-
Abstract: Tracking of cell movements in various cell culture setups is essential to many researchers in the life science sector. Gradientech AB, a Swedish biotech company, has developed CellDirector, a unique microfluidic system that academic researchers can use to study how concentration gradients of soluble proteins impact cell migration. The current project is focused on developing software for analyzing cell behavior and cell migration. The free open-source software CellProfiler developed at the Broad Institute will be used as a platform for a high-throughput system with automated high quality imaging, adapted for unlabeled cells, which are analyzed with regard to directionality of migration, speed, and acceleration. Apart from analyzing cell migration, the cell tracking aims at producing lineages, where cellular events such as cell division and cell death can be scored for single cells. A graphical user interface for visualizing and editing tracks imported from CellProfiler has been developed. This will be used for manual feed back in an iterative parameter optimization process, which aims to improve the automatic tracking. The progress of the project was presented in the poster session at eSSENCE Academy 2013 workshop at Lund.
Segmentation and Tracking of E.coli Bacteria in Bright-Field Microscopy Images
Sajith Kecheril Sadanandan, Carolina Wählby
Partners: Johan Elf and David Fange, Dept. of Cell & Molecular Biology, UU
Funding: SciLifeLab Uppsala, eSSENCE, VR junior researcher grant to CW
Period: 1210-
Abstract: Time-lapse microscopy is used to study the cellular and molecular processes in live cell experiments. Tracking of live cells and analysis of their spatiotemporal behavior is a common task in many experiments. This project aims to segment E.coli bacteria and to track them over time to construct the cell lineage. Bacteria are grown in a microfluidic device developed at Elf lab, Uppsala, which enables the imaging of monolayer cells. The unstained bright-field images of the cells are taken and a-priori information about the bacterial cells will be used to develop a system, which will have a GUI to set the parameters for proper segmentation and tracking. The results will be visually analyzed and the parameters are tuned. The optimized parameters will be used for the experiment to automatically analyze the data generated during the entire experiment.
Modelling Diffusion on Cell Surfaces
Ida-Maria Sintorn, Robin Strand
Partners: Ingela Parmryd, Dept. of Medical Cell Biology, UU; Jeremy Adler, Dept. Of Immunology, Genetics and Pathology, UU
Funding: TN-faculty, UU; S-faculty, SLU; VINNMER programme, Swedish Governmental Agency for Innovation Systems
Period: 1101-
Abstract: A cell surface is a highly irregular and rough. The surface
topography is however usually ignored in current models of the
plasma membrane, which are based on 2D observations of diffusion
that really occurs in 3D. In this project we model diffusion on
non-flat surfaces to explain biological processes occurring on the
cellsurface. During 2013, a poster was presented at Biophysical Society 2013 Annual Meeting, Philadelphia.
Analysis of Male Reproductive Tract Morphology in Reproductive Toxicology
Azadeh Fakhrzadeh, Cris Luengo, Gunilla Borgefors
Partners: Ellinor Spörndly-Nees, Lena Holm, Dept. of Anatomy, Physiology and Biochemistry, SLU
Funding: SLU (KoN)
Period: 1009-
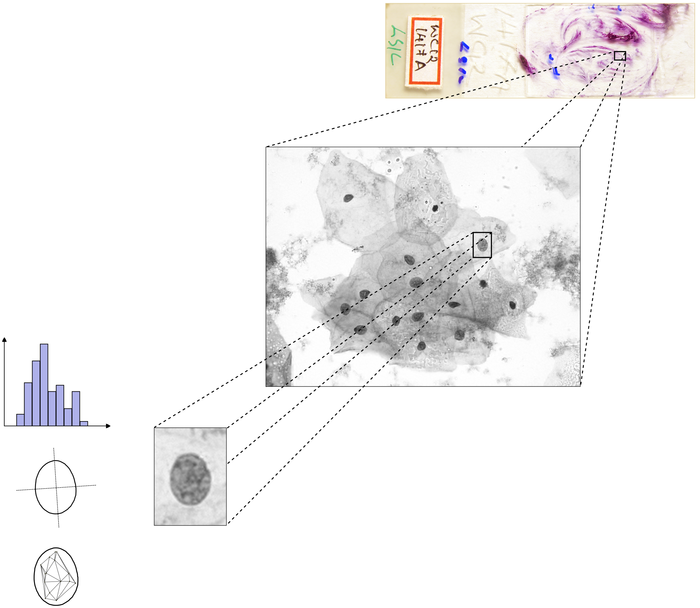
Abstract: Reproductive toxicology is the study of chemicals and their effects on the reproductive system of humans and animals. In reproductive toxicology, there is a strong need to detect structural differences in organs that often have both a complex microscopic structure and function. This problem is further complicated because standard techniques are based on the examination of two-dimensional sections of a three-dimensional structure. The aim of this project is to develop methods to objectively describe microscopic structures of male reproductive organs and to test these in reproductive toxicology research. The project is comparative and includes studies of organs from rooster and mink. We are developing automatic and interactive methods to analyze the relevant structures in the histology images of testis. We have constructed a semi-automatic method to delineate the epithelium cell layer in testicular tissue.
The cell nuclei are detected using the fast radial symmetry filter. A graph is constructed on top of the epithelial cells (Fig. 9). Graph-cut optimization method is used to cut the links between cells of different tubules. Generating sperms in seminiferous tubules is a cyclic process, during which various generations of germ cells in epithelial layer undergo a series of developmental steps. This cycle can be subdivided into 12 different stages. We are currently developing a texture-based classification method to determine each tubule's stage.
Automated Classification of Immunostaining Patterns in Breast Tissue from the Human Protein Atlas
Andreas Kårsnäs, Martin Simonsson, Carolina Wählby, Robin Strand
Partners: Caroline Kampf, The Human Protein Atlas (HPA); Virginie Uhlmann, Imaging Platform, Broad Institute of Harvard and MIT, Cambridge, Massachusetts MA, USA; S. Issac Niwas, P. Palanisamy, Dept. of ECE, National Institute of Technology (NIT), Tiruchirappalli, India
Funding: SciLifeLab Uppsala
Period: 1201-1303
Abstract: The Human Protein Atlas (HPA) is an effort to map the location of all human proteins (http://www.proteinatlas.org/) and contains a large number of histological images of sections from human tissue. Methods for quantification of staining patterns in histopathology have many applications, ranging from antibody quality control to tumor grading. In this project we have tested a new method based on complex wavelets textural features as well as an approach inspired by WNDCHARM (Weighted Neighbor Distances using a Compound Hierarchy of Algorithms Representing Morphology) for classifying nuclear versus cytoplasmic staining. During 2013, a paper was published in Journal of Pathology Informatics.
Combating Breast Cancer by Digital Pathology
Andreas Kårsnäs, Robin Strand, Carolina Wählby, Ewert Bengtsson
Partners: Visiopharm, Hørsholm, Denmark; Clinical Pathology Division, Vejle hospital, Vejle, Denmark
Funding: NordForsk Private Public Partnership PhD Programme and Visiopharm
Period: 0909-
Abstract: The results of analyses of tissue biopsies by pathologists are crucial for breast cancer patients. In particular, the precision of a patient's prognosis, and the ability to predict the consequences of various treatment opportunities before actually exposing the cancer patient, depend on the detection and quantification of biomarkers in tissue sections by microscopy. Experience from the last decade has revealed that manual detection and quantification of biomarkers by microscopy of tissue biopsies is highly dependent on the competencies and stamina of the individual pathologist. The aim of the present PhD project is to develop software-based algorithms that can facilitate the workflow and ensure objective and more precise results of the quantitative microscopy procedures in breast cancer.
During 2012, we worked on a project for verifying antibodies by comparing staining patterns in immune-stained histological images. The project was made in collaboration with the Human Protein Atlas project. We made a comparison of different methods for classifying staining patterns in histology. This work was presented at MICCAI'12 in Nice. We also presented the vectorial minimum barrier distance, a new method for computing gray-weighted distance transforms while incorporating vectorial data, at ICPR'12 in Tsukuba, Japan.
Early 2013, we started a new project aimed at developing a new method for registering histological images of consecutive sections with different staining. The project resulted in an article about multimodal registration using locally rigid transforms. The article is currently under review. In 2013, we also finished a journal article presenting a histopathological tool for sub-cellular quantification. The article was accepted early 2014 for publication in the journal Computer methods in Biomechanics and Biomedical Engineering: Imaging & Visualization.
Automatic, Quantitative Malignancy Grading of Prostate Cancer using Image Analysis
Ingrid Carlbom, Christophe Avenel
Partners: Christer Busch and Anna Tolf, Department of Immunology, Genetics and Pathology, University Hospital
Funding: The Swedish Research Council, Hillevi Fries Research Fund
Period: 1001-
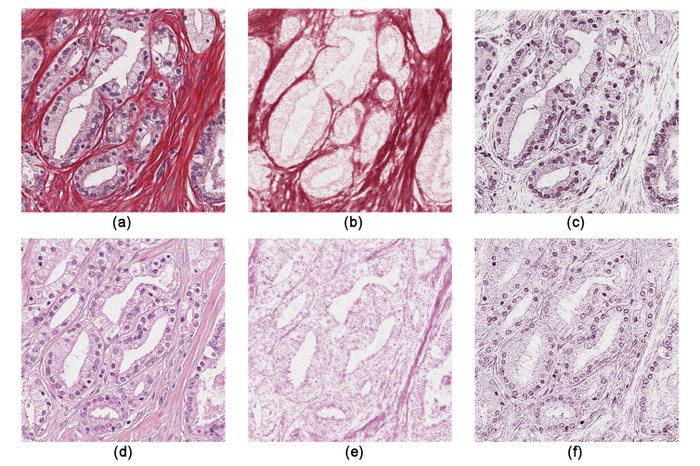
Abstract: Gleason grading is the most widely used system for determining the severity of prostate cancer. The Gleason grade is determined visually under a microscope from prostate tissue that is most often stained with Hematoxylin-Eosin (H&E).
|
Stain for blind color decomposition In an earlier study we demonstrated that H&E is not ideal for machine learning applications, but that other stains, such as Sirius-hematoxylin (Sir-Htx), may perform better. This year we demonstrated the advantages of this stain over H&E for blind color decomposition (Fig. 10). When compared to ground truth defined by an experienced pathologist, the relative root-mean-square errors of the color decomposition mixing matrices for Sir-Htx are better than those for H&E by a factor of two, and the Pearson correlation coefficients of the density maps resulting from the decomposition of Sir-Htx-stained tissue gives a 99% correlation with the ground truth. Qualitative examples of the density maps confirm the quantitative findings and illustrate that the density maps will allow accurate segmentation of morphological features that determine the Gleason grade. Identification of epithelial nuclei From the epithelial density map, resulting from the blind color decomposition of Sir-Htx-stained prostate tissue, we used a marked point process to segment the epithelial nuclei (Fig. 11). This enables us to extract nuclei as individual, joint, or overlapping objects generally without discarding overlapping parts and therefore without major loss in segmentation precision. The algorithm, which was originally developed for breast cancer tissue nuclei identification, uses simulated annealing combined with a "birth and death" process to find the best match with the density map, and was adapted to prostate tissue by pre-and-post processing methods.
Database of images from whole mount sections We have created two online tools in order to build a database of graded images. The image selection tool is based on OpenSeaDragon (an open-source, web-based viewer for zoomable images) and facilitates the selection of small images from whole mount sections. With this tool we are building an image database where each image has a dominant pattern that represents for example one Gleason grade, benign tissue, stroma, or artifacts such as a tear in the tissue. The grading tool allows multiple pathologists to grade and comment on the previously selected images, without seeing each other grades, and is a basis for a consensus-graded database for developing and testing automatic Gleason grading algorithms (Fig. 12).
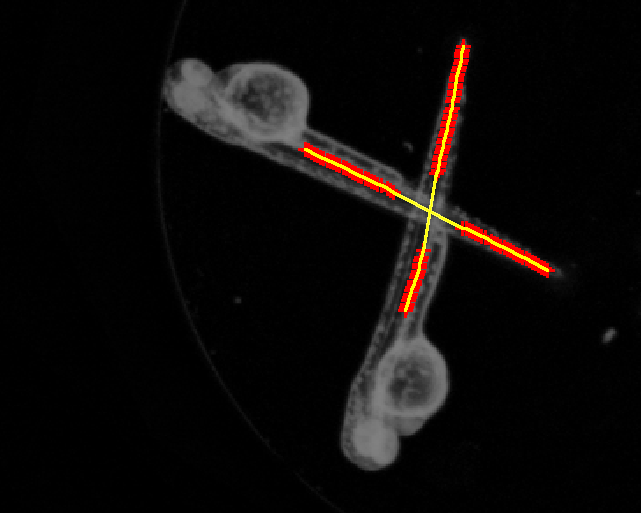
Automated Quantification of Zebrafish Tail Deformation for High-throughput Drug Screening
Omer Ishaq, Alexandra Pacureanu, Carolina Wählby
Partners: Joseph Negri, Mark-Anthony Bray, Randall T. Peterson, Broad Institute of Harvard and MIT
Funding: SciLifeLab Uppsala
Period: 1203-1304
Abstract: Zebrafish (Danio rerio) is an important model organism in biomedical research due to its ease of handling and translucent body and consequently many human disease models have been established in the Zebrafish. Zebrafish embryos undergo spinal deformation upon exposure to chemical agents, such as Camptothecin (Cpt), that inhibit DNA repair. We are developing automated image-based quantification of spine deformation enabling whole-organism based assays for use in early-phase drug discovery campaigns. Our automated method for accurate high-throughput measurement of tail deformations in multi-fish micro-plate wells generates refined medial representations of partial tail-segments. Subsequently, these disjoint segments are analyzed and fused to generate complete Zebrafish tails (Fig. 13). Based on these estimated tail curvatures we reach a classification accuracy of 91% on individual animals as compared to known control treatment. This accuracy is increased to 95% when combining scores for fish in the same well. A paper describing the methods and results was published and presented at the International Symposium for Biomedical Imaging (ISBI) in April 2013.
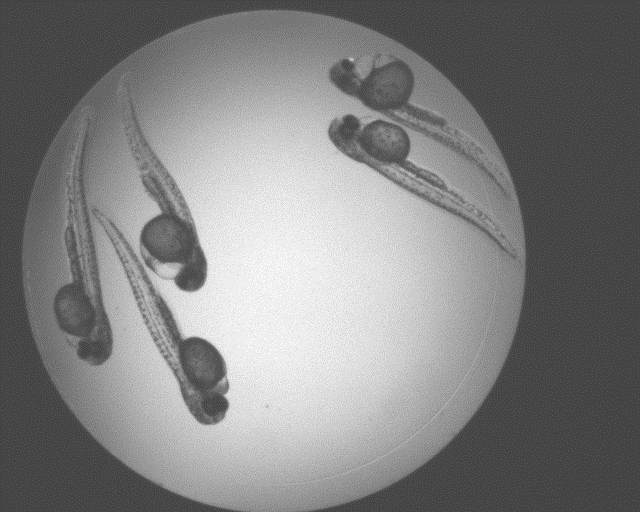
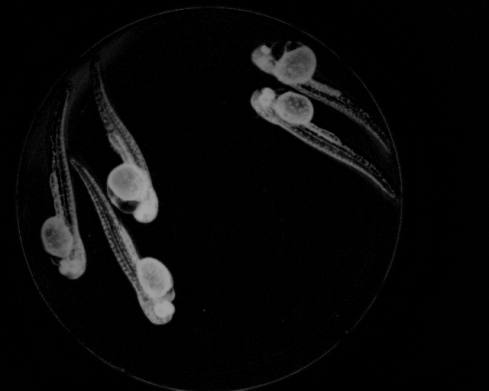
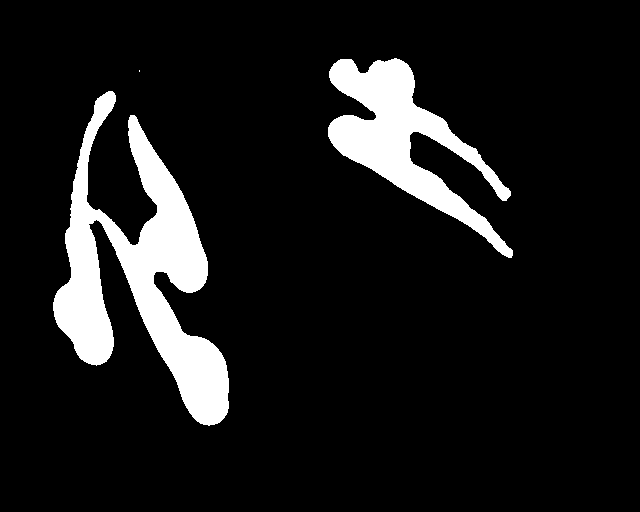
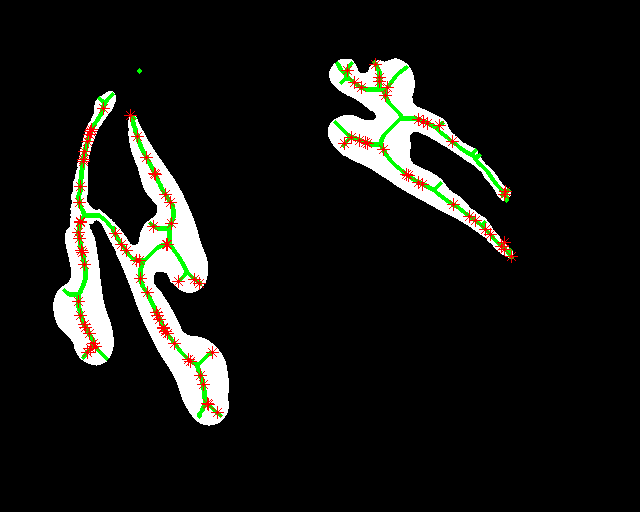

|
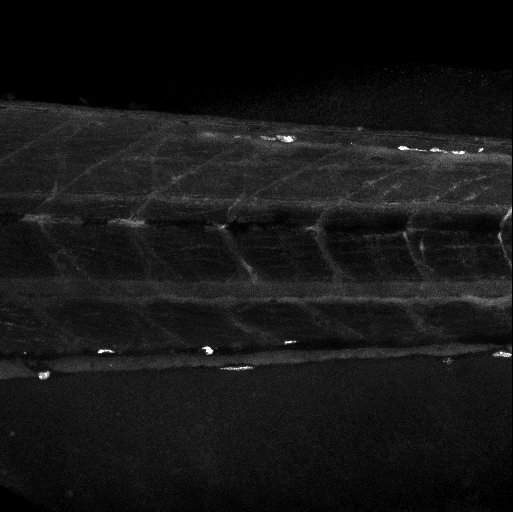
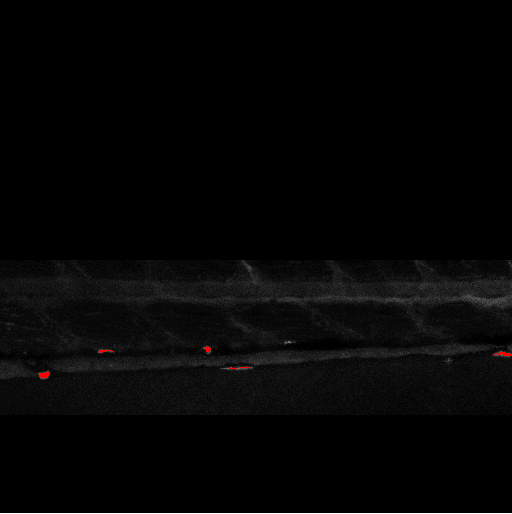
Quantification of Zebrafish Lipid Droplets
Petter Ranefall, Carolina Wählby
Partners: Marcel den Hoed, Manoj Bandaru, Erik Ingelsson, Department of Medical Sciences and SciLifeLab, UU
Funding: SciLifeLab Uppsala
Period: 1308-
Abstract: The aim of this project is to identify novel targets for the therapeutic intervention of coronary artery disease. This is done by following-up results from genome-wide association studies in epidemiological studies using a zebrafish model system. Using image analysis we try to identify and characterize causal genes within loci that have so far been identified as associated with coronary heart disease by (high-throughput) screening of atherogenic processes in wildtype and mutant zebrafish, both before and after feeding on a control diet or a diet high in cholesterol. Using confocal microscopy we can image fat accumulation in the zebrafish (Fig. 14).
|
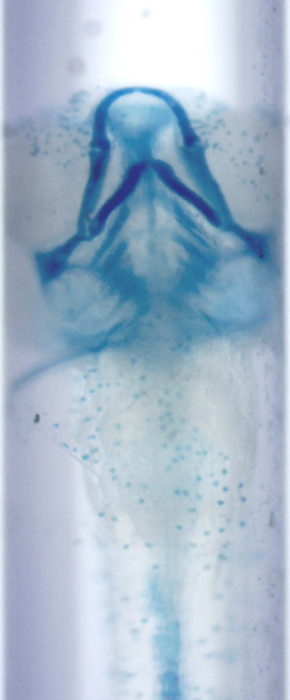
Optical Projection Tomography
Alexandra Pacureanu, Omer Ishaq, Carolina Wählby
Partners: Amin Allalou, Izolde AB, Uppsala; Johan Ledin, Evolutionary Biology Centre, Zebrafish platform, SciLifeLab Uppsala; Jos Buijs, Ridgeview Uppsala, Carlos Pardo, Mehmet F. Yanik, Research Laboratory of Electronics, Massachusetts Institute of Technology, Cambridge, USA
Funding: SciLifeLab Uppsala; TN-faculty, UU
Period: 1009-
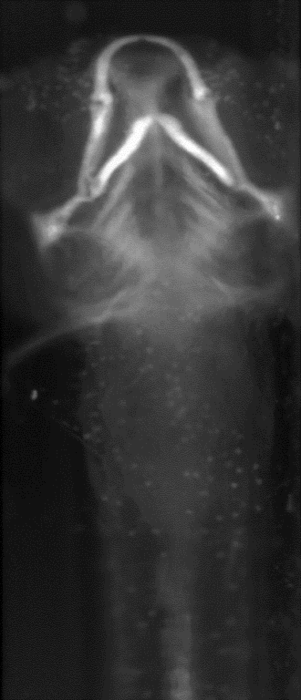
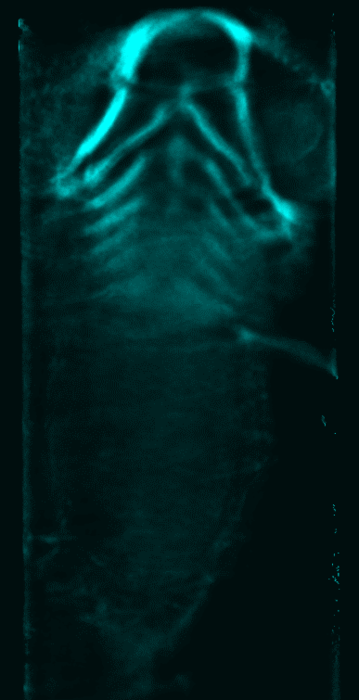
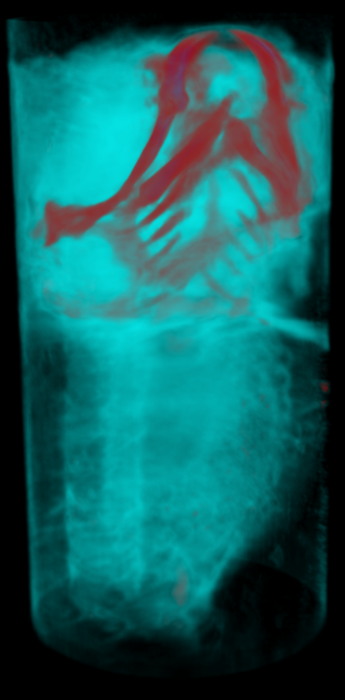
Abstract: Isotropic 3D imaging of biological specimens is instrumental for further breakthroughs in life sciences. Many biological specimens with high relevance for basic research, disease studies and drug discovery, such as model organisms or 3D cell cultures, are semi-transparent to visible light. This lead to the advent of the technique dubbed optical projection tomography (OPT). The 3D internal structure is revealed by the attenuation variations of the light traversing the specimen. In OPT transverse slices of the specimen are reconstructed from a set of angular projections and stacked together into a volumetric image. This method enables in vivo imaging of relatively large samples with high spatial resolution. A high-throughput platform for cellular resolution, in vivo OPT of zebrafish has been developed at MIT, Cambridge, USA. With this system we have shown that OPT of zebrafish embryos can provide 3D information enabling high-throughput screening of subtle phenotypic changes in relation to drug treatment, as published in Nature Communications in February 2013. However, OPT imaging systems in general are still quite sophisticated and costly. We are therefore developing a system for optical 3D isotropic imaging at microscopic scale, based on readily accessible hardware. The total price of the setup is kept under 1000 euros and the components can be easily obtained around the world. We have assembled the image acquisition system, acquired, and reconstructed images of zebrafish embryos (Fig. 15) and of 3D cell cultures (Fig. 16). We are complementing the simple hardware with open source computational tools, embedding algorithms for image alignment, correction and reconstruction. Our goal is to enable every life sciences research laboratory to have access to valuable 3D information on biological specimens.
In 2013, besides working on improving imaging of zebrafish embryos, we attempted to image 3D cell cultures with our system, in collaboration with Jos Buijs (Ridgeview). A human ovarian carcinoma cell line has been used to grow 3D cell cultures in borosilicate thin tubes. We also tested growing the cells in agar gels and performing a 'biopsy' to extract the cells and transfer them into borosilicate tubes for imaging. We presented a poster at IEEE International Symposium on Biomedical Imaging (ISBI), San Francisco, USA and a journal manuscript is under preparation.
|
Image-based Approaches for Drug Tablet Quality Assessment
Ida-Maria Sintorn, Carolina Wählby
Partners: Mark Nicholas, Mats Josefson, AstraZeneca, Mölndal, Sweden
Funding: Pre-study grant from AIMDay Image, UU Innovation
Period: 1204-1302
Abstract: It is known qualitatively that microstructural differences in solid dosage forms (e.g. tablets and inhalation powders) affect the performance of the medication. The microstructural differences are differences in the spatial distribution of active and inactive compounds. The aim of this project is to characterize these microstructural differences in order to determine whether imaging techniques such as CLSM (confocal laser scanning microscopy), wide-field fluorescence microscopy, and TOF-SIMS (Time-Of-Flight Secondary Ion Mass Spectroscopy) can reveal quantifiable differences in structure. The problem was addressed using a combination of local intensity features and texture measurements (including granulometry, Zernike moments, and Haralick features), and measurements were correlated with tablet characteristics/treatments. Due to a relatively limited dataset, it was difficult to find statistically significant differences. The data was presented to AstraZeneca researchers in January 2013.
Tools for Analysis and Visualization of Giga-Pixel Sized Slide-Scanner Images.
Petter Ranefall, Alexandra Pacureanu, Carolina Wählby
Partners: Mats Nilsson, Thomas Hauling, Marco Mignardi, Jessica Svedlund, Elin Lundin, Department of Biochemistry and Biophysics and SciLifeLab, Stockholm University.
Funding: SciLifeLab
Period: 1308-
Abstract: The aim is to create a tool for full resolution image analysis of large images, e.g. slide scanner data, with the possibility of visual examination and interaction at multiple resolutions. The tool is built on a free and open-source framework for visual examination at multiple resolutions with the option to toggle results on or off, such as segmentation masks, classification results, and tissue morphology measurements, using a map view with seamless zooming and panning capabilities, allowing for fast navigation between a full-tissue view and high-resolution sub-cellular observations (Fig. 17). The aim is to also have an interface that enables visual/manual selection of regions of interest, target discovery, and understanding of novel spatial relationships within the tissue environment.