Elisabeth Linnér, Robin Strand
Partner: Joel Kullberg, Dept. of Radiology, Oncology and Radiation Science, UU
Funding: TN-faculty, UU
Period: 1005-
Abstract: Most three-dimensional image acquisition methods are based on tomography, i.e., the three-dimensional image is constructed by a stack of two-dimensional images, each acquired separately. To take full advantage of the high sampling efficiency of the optimal lattices, the image should be acquired as a full three-dimensional image. This is possible in magnetic resonance imaging. In this project, methods for image acquisition, analysis, and visualization will be developed for magnetic resonance images represented on non-standard lattices.
With the methods developed in this project, it will be possible to process three dimensional images with 30% less data without affecting the image quality. Our intention is that this project will lead to faster processing of images with less demands on the data storage capacity.
Robiël Naziroglu, Catherine Östlund, Cris Luengo
Partner: Innventia AB, Stockholm
Funding: VINNMER programme, Swedish Governmental Agency for Innovation Systems
Period: 1008-1012
Abstract: A new table-top micro-CT device was installed at Innventia in 2010. We studied the device's noise characteristics, the relationship between the noise in the raw data and that in the reconstructed volumetric data, the effect on the image of different exposure times, and the use of filters during acquisition. We developed a method to accurately estimate the local volume fraction of fibre in images of paper and cardboard. We also investigated the correlation between noise in neighbouring voxels in these images, and developed a method to more accurately classify voxels as fibre or air using these statistics.
Khalid Niazi, Ingela Nyström
Partners: M. Talal Ibrahim, Ling Guan, Ryerson Multimedia Lab, Ryerson University, Canada
Funding: COMSATS IIT, Islamabad
Period: 1003-
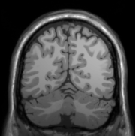
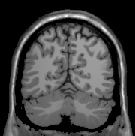
Abstract: Non-uniform illumination is considered as one of the major challenges in the field of medical imaging. It is often caused by the imperfections of the data acquisition device and the properties of the object under study. We have developed an iterative method which suppresses the magnitude of the frequencies that are responsible for non-uniformity in an image using the gray-weighted distance transform (GWDT). Moreover, the proposed method is not user dependent as all the parameters are automatically generated on the basis of the GWDT. It is tested on images acquired from several imaging modalities which makes it different and unique from most of the existing methods. Fig.
|
[]  []
[] 
|
Ewert Bengtsson, Ingela Nyström
Partners: Stuart Crozier, Andrew Mehnert, School of Information Technology and Electrical Engineering, University of Queensland, Brisbane, Australia, MedTech West
Funding: TN-faculty, UU; The Australian Research Council
Period: 0503-
Abstract: The pattern of change of signal intensity over time in dynamic contrast enhanced magnetic resonance images (DCE-MRI) of the breast is an important criterion for the differentiation of malignant from benign lesions. Malignant lesions release angiogenic factors which induce the growth and sprouting of existing blood vessels and the formation of new leaky vessels. This gives rise to increased inflow and an accelerated extra-vascularisation of contrast agent at the tumour site which is reflected as T1-weighted signal increase. However strong enhancement is not specific to malignant lesions. Contrariwise shallow or no enhancement is a feature of some malignant lesions. As a result the specificity of the technique is poor to moderate.
This project is seeking to improve the specificity of breast MRI, and therefore its clinical utility, in two ways. The first is by combining features describing enhancement with those describing lesion morphology and micro-structure. This involves integrating intrinsic contrast MRI with DCE-MRI and diffusion-weighted (DW) MRI. The second is by reducing subjectivity in routine clinical interpretation of breast MRI data by means of computer visualisation, image analysis, and statistical pattern recognition.
This collaborative project has evolved from one which Bengtsson joined during his sabbatical at the University of Queensland in 2004-2005. Significant outcomes to date include novel software and algorithms for motion correction and registration, registration evaluation, denoising, parametric modelling of contrast enhancement, 3D colour-coding of 4D DCE-MRI data, hardware-accelerated volume visualisation using a colour-preserving maximum intensity projection in the HSV colour space, haptic interaction/interrogation of the volumetric data, breast and lesion segmentation, feature (morphology, enhancement, and diffusion) extraction, and lesion classification. The project has also developed several plug-ins for the open source OsiriX platform specifically for visualisation, reporting/annotation, and image analysis/processing of breast MRI data. This software is currently being evaluated under license by a private radiology practice in Queensland.
Collaboration is continuing on the evaluation of the developed parametric models of enhancement as well as the development of DW-MRI features to characterise micro-structural changes in malignant tissue.
Lennart Svensson, Ida-Maria Sintorn, Stina Svensson, Ingela Nyström, Anders Brun, Ewert Bengtsson
Partners: Dept. of Cell and Molecular Biology, Karolinska Institute; SenseGraphics AB
Funding: The Visualization Program by Knowledge Foundation; Vaardal Foundation; Foundation for Strategic Research; Swedish Governmental Agency for Innovation Systems; Invest in Sweden Agency
Period: 0807-
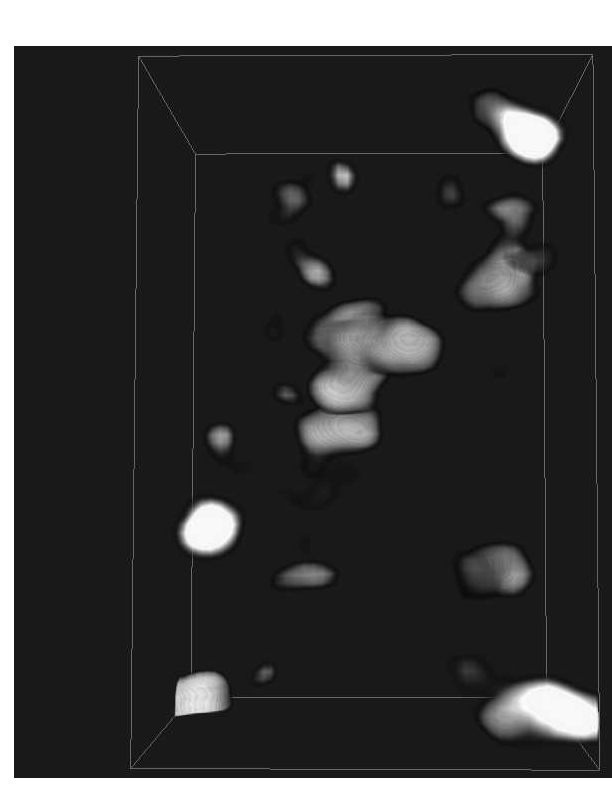
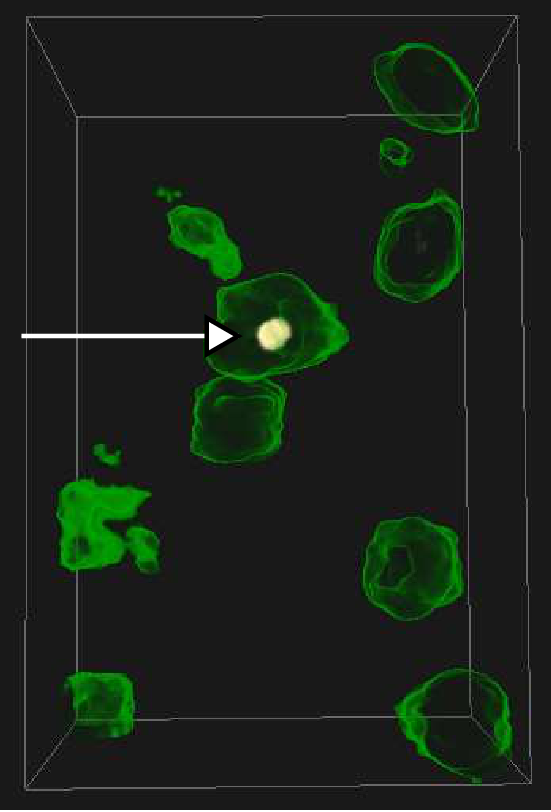
Abstract: The traditional methods for solving the structure of proteins are X-ray crystallography and NMR spectroscopy. An alternative approach, Molecular Electron Tomography (MET), has more recently gained interest within the field of structural biology as it enables studies of individual structures in tissue at nanometer scale, whereas the old methods only allowed studies in solution. Additionally, MET enables studies of the dynamics of proteins. MET results in images of low resolution, as compared with e.g., X-ray crystallography, and low signal-to-noise ratio. A new kind of software for postprocessing is hence required, which allows for proper visualization and analysis, as well as data fusion with measurements from X-ray crystallography and NMR. The target of the ProViz project is to create these software tools. Our approach is to use stereo visualization and haptic rendering of the images in order to facilitate a good understanding of the data as well as interactivity in semi-automatic methods.
During 2010, we have focused on developing
methods for automatic transfer function generation for volume
rendering of MET data-sets. A paper investigating different image
properties for automatically generating a suitable transfer function
will be presented at the International conference on Computer
Graphics, Visualization and Computer vision early 2011. We have also
investigated methods for visualizing the goodness of fit when matching
or docking other types of data (e.g. X-ray crystallography data) in
the MET volumes. See example in Fig. ![]() .
.

|
Filip Malmberg, Ingela Nyström, Ewert Bengtsson, Stefan Seipel
Partners: Gunnar Jansson, Dept. of Psychology, UU;
Funding: Swedish Research Council; TN-faculty, UU
Period: 0301-
Abstract: Modern medical imaging techniques provide 3D images of increasing complexity. Better ways of exploring these images for diagnostic and treatment planning purposes are needed. Combined stereoscopic and haptic display of the images form a powerful platform for such image analysis. In order to work with specific patient cases, it is necessary to be able to work directly with the medical image volume and to generate the relevant 3D structures as they are needed for the visualization. Most work so far on haptic display use predefined object surface models. In this project, we are creating the tools necessary for effective interactive exploration of complex medical image volumes for diagnostic or treatment planning purposes through combined use of haptic and 3D stereoscopic display techniques. The developed methods are tested on real medical application data. Our current applications are described further in Projects
A software package for interactive visualization and segmentation developed within this project has been released under an open-source license. The package, called WISH, is available for download at http://www.cb.uu.se/research/haptics.
Filip Malmberg, Ewert Bengtsson, Ingela Nyström
Partners: Andrew Mehnert and Craig Engstrom, ITEE Dept., University of Queensland, Brisbane, Australia
Funding: TN-faculty, UU; The Australian Research Council
Period: 0803-
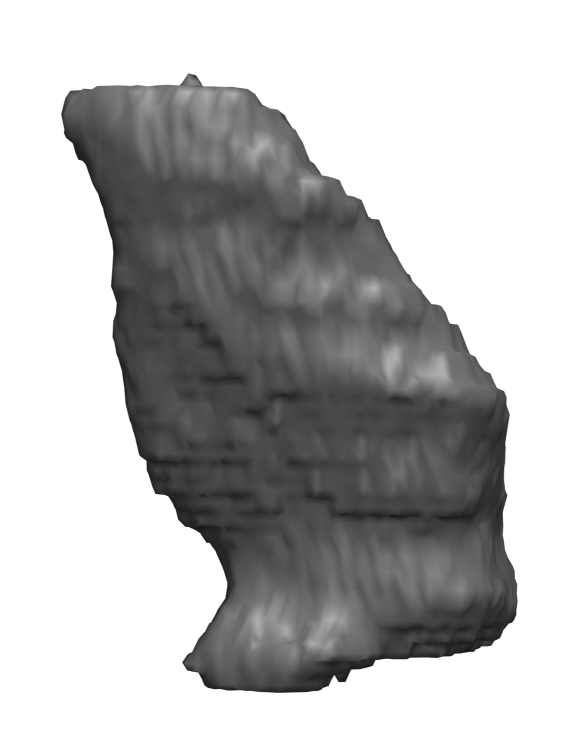
Abstract: In cricket fast bowlers, substantial volume asymmetries of the quadratus lumborum (QL) muscle have been associated with a significantly increased risk of developing lumbar spine stress fractures. In this project, we aim to measure such asymmetries by segmenting the QL muscle in MR images. At the University of Queensland, an automatic segmentation method has been developed for this purpose. The method uses a shape atlas, built from a large number of reference segmentations, to segment a specific muscle. Currently, we are investigating if interactive segmentation methods can be used to accelerate the creation of reference segmentations. By accelerating this process, we make it easier to adapt the atlas-based automatic segmentation method to other interesting muscles. A 3D visualization of a segmented QL muscle is shown in Fig.
Ingrid Carlbom, Ewert Bengtsson, Filip Malmberg, Ingela Nyström, Stefan Seipel, Pontus Olsson, Robin Strand, Martin Ericsson, Fredrik Nysjö
Partners: Stefan Johansson (Teknovest AB); Jonny Gustafsson and Lars Mattson, Industrial Metrology and Optics Group, Royal Institute of Technology; Jan-Michaél Hirsch, Dept. of Surgical Sciences, Oral & Maxillofacial Surgery, UU, Consultant at Dept. of Plastic- and Maxillofacial Surgery, UU Hospital; Roland Johansson, Dept. of Neurophysiology, Umeå University; Håkan Lanshammar, Kjartan Halvorsen, Dept. of Information Technology, UU; PiezoMotors AB, SenseGraphics AB
Funding: Knowledge Foundation
Period: 0908-
Abstract: We are building an augmented reality system that will allow users to touch and manipulate high contrast, high resolution, three-dimensional virtual objects suspended in space using a glove that gives realistic whole hand haptic feedback both in terms of force feedback to the wrist and finger joints and tactile feedback to the fingertips. The driving application is cranio-maxillofacial surgery, giving a surgeon the ability to plan complex procedures, moving bone and implants into their desired positions and then stretching the soft tissue to accommodate the structural changes. However, the system may also significantly enhance other applications in, for example, computer-aided design, training simulators for object assembly, and 3D games.
During 2010 we assembled the first generation display system with a Holographic Optical Element and evaluated its rendering fidelity, latency, and perceived 3D-accuracy, resulting in several new design criteria for the next generation display, including a more optimal separation of the adjacent views. Perspective correction (keystone correction) warps the images from the projectors so that they coincide, and a commercial head tracker enables removal of the discontinuities between adjacent views. We also designed a series of interaction experiments to evaluate the feasibility of co-located haptic interaction with the Holographic Optical Element.
The first generation haptic glove acts as an external skeleton with the hand and finger joints controlled by actuators that are integrated into the glove (Fig. ![]() ). This first prototype has six degrees of freedom (DOF) movement of the hand and one DOF gripping with the thumb and index finger. The six DOF movements are accomplished with a commercial haptic arm, the SensAble Phantom Premium, while the gripping haptic glove is developed within this project. The gripping force is controlled by the most compact, high-precision piezoelectric motor that is commercially available today, a piezoelectric motor from Piezomotor AB, using a separate force sensor in a feed-back loop. The high stiffness of the motor in combination with a very high dynamic speed range allows for delicate grip force control. This first single joint glove has identified the most important issues for subsequent generations: the actuators have to be further miniaturized with the speed increased, the audible sound eliminated, and the weight reduced.
). This first prototype has six degrees of freedom (DOF) movement of the hand and one DOF gripping with the thumb and index finger. The six DOF movements are accomplished with a commercial haptic arm, the SensAble Phantom Premium, while the gripping haptic glove is developed within this project. The gripping force is controlled by the most compact, high-precision piezoelectric motor that is commercially available today, a piezoelectric motor from Piezomotor AB, using a separate force sensor in a feed-back loop. The high stiffness of the motor in combination with a very high dynamic speed range allows for delicate grip force control. This first single joint glove has identified the most important issues for subsequent generations: the actuators have to be further miniaturized with the speed increased, the audible sound eliminated, and the weight reduced.
We built a first tactile array prototype comprising a single array 1.4 x 1.4 mm element, intended for an array with 2 mm pitch. Tests show that this type of solution can be used in array formats and that each element can generate lateral forces. The lateral speed is about 5 mm/sec that is close to the final requirement. It is also possible to move objects with very small mass that is required for skin stimuli. The design is based on piezoelectric bimorphs that can be surface mounted on flexible printed circuit boards, i.e., the design is already prepared for industrial production. The ultimate solution will require further development to fulfill all requirements.

|
Filip Malmberg, Ewert Bengtsson, Ingela Nyström
Partners: Jan Michael Hirsch, Elias Messo, Dept. of Surgical Sciences, UU Hospital
Funding: TN-faculty, UU; NovaMedTech
Period: 0912-
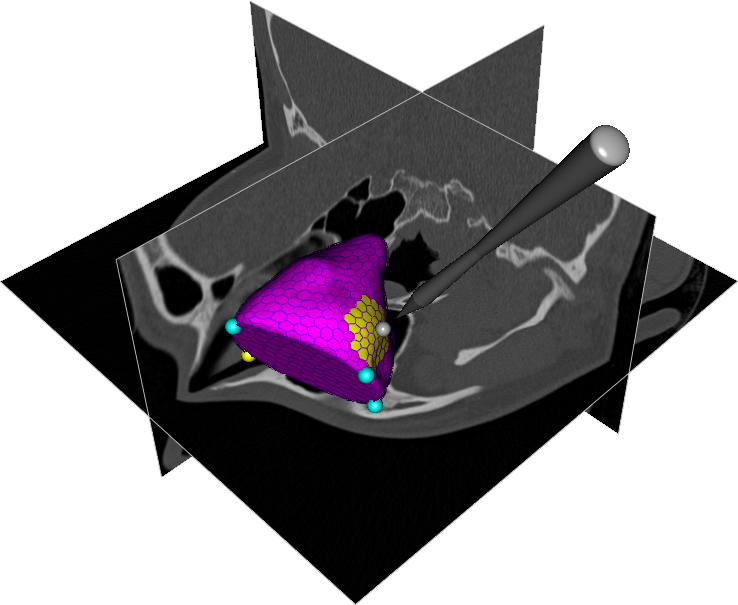
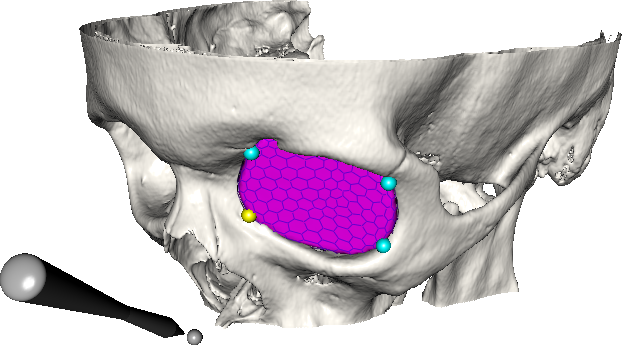
Abstract: A central problem in cranio-maxillofacial (CMF) surgery is to restore the normal anatomy of the skeleton after defects, i.e., malformations, tumors and trauma to the face. This is particularly difficult when a fracture causes vital anatomical structures such as the bone segments to be displaced significantly from their proper position, when bone segments are missing, or when a bone segment is located in such a position that any attempt to restore it into its original position poses considerable risk for causing further damage to vital anatomical structures such as the eye or the central nervous system. There is ample evidence that careful pre-operative planning can significantly improve the precision and predictability and reduce morbidity of the craniofacial reconstruction. In addition, time in the operating room can be reduced. An important component in surgery planning is to be able to accurately measure the extent of certain anatomical structures. Of particular interest in CMF surgery are the shape and volume of the orbits (eye sockets) comparing the left side with the right side. These properties can be measured in CT images of the skull, but this requires accurate segmentation of the orbits. Today, segmentation is usually performed by manual tracing of the orbit in a large number of slices of the CT image. This task is very time-consuming, and sensitive to operator errors. Semi-automatic segmentation methods could reduce the required operator time significantly. In this project, we are developing a prototype of a semi-automatic system for segmenting the orbit in CT images. Segmentations obtained with this system are shown in Fig.
|
Filip Malmberg, Ingela Nyström, Ewert Bengtsson
Funding: TN-faculty, UU
Period: 0901-
Abstract: Image segmentation, the process of identifying and separating relevant objects and structures in an image, is a fundamental problem in image analysis. Accurate segmentation of objects of interest is often required before further processing and analysis can be performed. Despite years of active research, fully automatic segmentation of arbitrary images remains an unsolved problem.
Seeded segmentation methods attempt to solve the segmentation problem in the presence of prior knowledge in the form of a partial segmentation. Given an image where a small subset of the image elements (called seed-points) have been assigned correct segmentation labels (e.g., object or background), an automatic algorithm completes the labeling for all image elements. The seed-points may be provided either by some automatic pre-processing algorithm, or by a human user in an interactive setting.
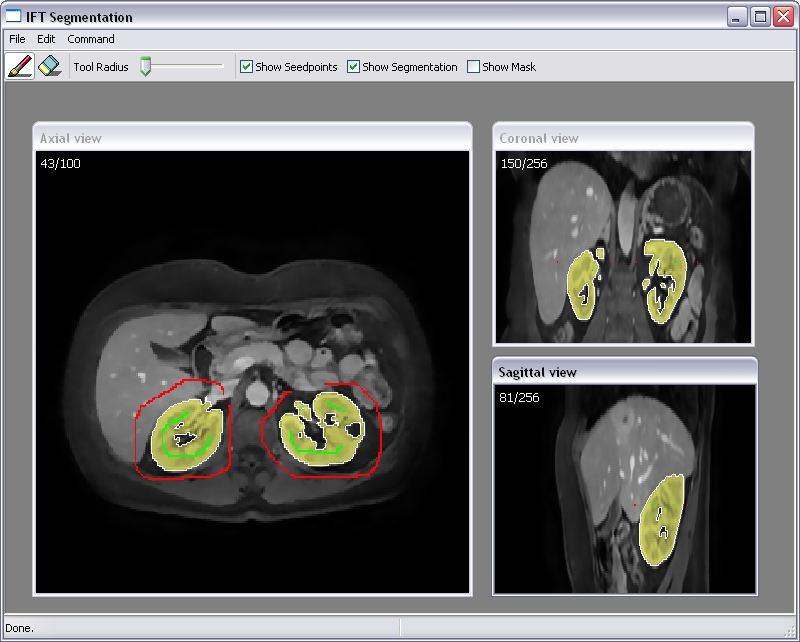
This project concerns the development of general tools for interactive seeded segmentation. In particular, we are studying methods based on the Image Foresting Transform (IFT). A method for incorporating smoothness into the IFT framework was presented at the SPIE Medical Imaging 2010 conference. An illustration of a segmentation obtained with this method is shown in Fig. ![]() . A theoretical framework for performing graph-based segmentation with sub-pixel precision has also been developed, in close collaboration with Project
. A theoretical framework for performing graph-based segmentation with sub-pixel precision has also been developed, in close collaboration with Project ![]() .
.
Filip Malmberg, Ingela Nyström, Ewert Bengtsson
Partners: Sven Nilsson, Milan Golubovic, Dept. of Oncology, Radiology, and Clinical Immunology, UU Hospital
Funding: Swedish Research Council; TN-faculty, UU
Period: 0501-
Abstract: The manual step in semi-automatic segmentation of medical volume images typically involves initialization procedures, such as placement of seed-points or positioning of surface models inside the object to be segmented. The initialization is then used as input to an automatic segmentation algorithm. We investigate how such initialization tasks can be facilitated by using haptic feedback. In this project, we develop interactive methods for segmenting the organs from abdominal CT scans. For example, liver segmentation is of importance in hepatic surgery planning, where it is a first step in the process of finding vessels and tumours, and the classification of liver segments. Liver segmentation may also be useful for monitoring patients with liver metastases, where disease progress is correlated to enlargement of the liver. We have developed a fully 3D liver segmentation method where high accuracy and precision is efficiently obtained via haptic interaction in a 3D user interface. Our method makes it possible to avoid time-consuming manual delineation, which otherwise is a common option prior to, e.g., hepatic surgery planning. Currently, we are incorporating and adapting region-based segmentation methods, such as the image foresting transform (IFT), into our software. Fig.
Ewert Bengtsson, Anders Hast
Partner: Tony Barrera, Uppsala
Funding: TN-faculty, UU
Period: 9911-
Abstract: Computer graphics is increasingly being used to create realistic images of 3D objects for applications in entertainment, (animated films, games), commerce (showing 3D images of products on the web), industrial design and medicine. For the images to look realistic high quality shading and surface texture and topology rendering is necessary. A proper understanding of the mathematics behind the algorithms can make a big difference in rendering quality as well as speed. We are in this project re-examining some of the established algorithms and are finding new mathematical ways of simplifying the expressions and increasing the implementation speeds without sacrificing image quality. We have also invented a number of completely new algorithms. The project is carried out in close collaboration with Tony Barrera, an autodidact mathematician. It has been running since 1999 and resulted in more than 25 international publications and a PhD thesis.
Ewert Bengtsson, Patrik Malm, Hyun-Ju Choi, Bo Nordin
Funding: Swedish Governmental Agency for Innovation Systems (Vinnova) and Swedish Research Council
Period: 0801-
Abstract: Cervical cancer is killing a quarter million women every year. Screening based on so called PAP-smears have proven very effective to reduce cancer mortality but require much work of well trained cytotechnologist. For 50 years research to automate the screening has been in progress, Bengtsson was very active in this field 1973-1993. Since about 10 years, commercial automated systems have been in operation but unfortunately those systems have many limitations.
In India there is no effective screening program in operation and around 70,000 women die from the disease each year. Now an effort to develop a screening system adapted to Indian situation is in progress at the research institute CDAC in Thiruvananthapuram, Kerala in cooperation with the Regional Cancer Centre there. Based on our earlier experience in the field we are cooperating with this project and we have received support from Vinnova and VR for this. We are in particular studying whether the 3D chromatin texture of the cervical cells can be utilized as a robust feature for detecting (pre-) cancerous lesions.
So far we have developed a new approach to segmentation and a method for generating synthesized PAP-smear images which can be used for testing the performance of new segmentation methods, the latter presented at ISBI 2010.
At CDAC they have developed an overall framework for the screening and a program for data collection, they are currently working on improved segmentation and feature extraction methods.
In September 2010 a delegation from CDAC and RCC visited CBA for a workshop where we in detail discussed the progress in the project. That was followed by a visit of Bengtsson to India in late December 2010- early January 2011. During that visit the plans for the continued work on the project were updated.
Dr Hyun-Ju Choi from Korea has studied 3D nuclear texture for
other applications and she was a visiting researcher at CBA
November 2008 to May 2010, see also Project ![]() .
During that stay she developed texture analysis methods that will be used in this project.
.
During that stay she developed texture analysis methods that will be used in this project.
Hyun-Ju Choi, Ewert Bengtsson
Funding: Korea Research Foundation and Swedish Research Council
Period: 0811-1005
Abstract: Hyun-Ju Choi was a visiting researcher at CBA 2008-2010. During this time she is developing 3D texture analysis methods. The standard 2D texture analysis methods, GLCM and GLRLM, have been extended and implemented for 3D volume data. New feature sets based on fractal analysis and wavelet transform for describing quantitatively nuclear chromatin changes in 3 dimensions have been developed. The measures have been tested on different data sets and after her return to Korea in May 2010 she has continued working with 3D texture analysis. The plan for the near future is to use the developed methods with image stacks from PAP-smears to detect malignancy related changes.