Robin Strand, Ingela Nyström, Gunilla Borgefors, Stina Svensson
Funding: UU TN Faculty, Graduate School in Mathematics and Computing (FMB), SLU S Faculty
Period: 9501-
Partners: Gabriella Sanniti di Baja, Istituto di Cibernetica, CNR, Pozzuoli, Italy; David Brunner, Chemnitz University of Technology, Chemnitz, Germany
Abstract: Skeletonization is a way to reduce dimensionality of digital objects. A skeleton should have the following properties: topologically correct, centred within the object, thin, and fully reversible. In general, the skeleton can not be both thin and fully reversible. We have been working on 3D skeletonization for the last decade.
Topology preservation is guaranteed by removing only simple points. Usually, a condition based on the number of connected components in a small neighbourhood is considered to test whether a grid point is simple or not. Such a condition for grid points on the body-centered cubic (bcc) grid is presented and proven to be correct in the report Simple points on the body-centered grid, see 6.5.7. In the report, another condition for directional thinning is also proven to be correct. The condition is used to develop a directional thinning approach in A high-performance parallel thinning approach using a non-cubic grid structure, see 6.5.8. The resulting skeleton is a curve skeleton that is thin, but not reversible.
Robin Strand, Céline Fouard, Gunilla Borgefors, Stina Svensson
Funding: SLU S Faculty, Graduate School in Mathematics and Computing (FMB)
Period: 9309-
Partner: Benedek Nagy, Dept. of Computer Science, Faculty of Informatics, University of Debrecen, Debrecen, Hungary
Abstract: The distance between any two grid points in a grid is defined by a distance function. The distance functions considered in this project (in contrast to Project 3) only depend on the positions of the grid points. During 2006, two kinds of path-generated distance functions are considered in this project. For path-generated distance functions, the distance between two points is defined as the shortest path between the points. To define paths between points, an adjacency relation and the cost (weight) for a step between two neighbouring grid points must be defined. The rotational dependency can be minimized either by using predefined weights (weighted distances) or by varying the adjacency relation along the path (distance based on neighbourhood sequences).
By combining weighted distances with distance based on neighbourhood sequences, a distance function with very low rotational dependency is obtained. Some theoretical results about such distance functions have been derived in two manuscript submitted for publication.
Weighted distance functions and distance transforms have been examined in a very general framework -- modules and point-lattices, respectively. The paper describing these results has been accepted for publication in Pattern Recognition.
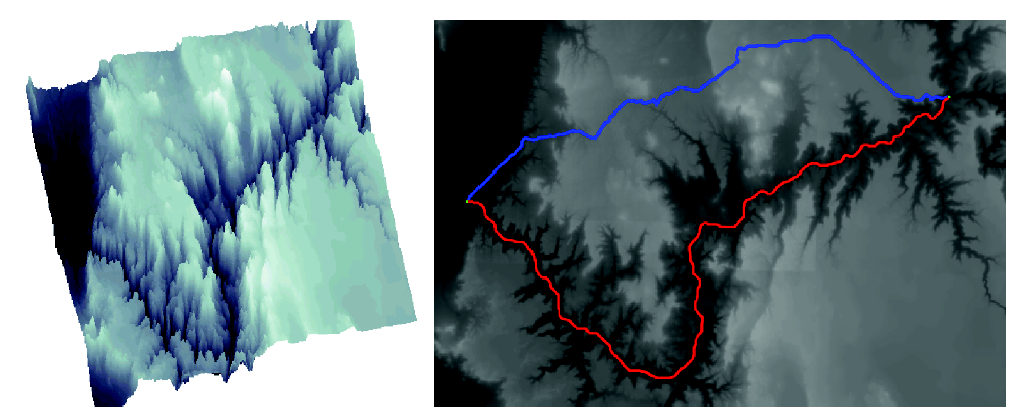
Distances based on neighbourhood sequences on the fcc and bcc grids, see Project 4, have also been examined. This research has resulted in a number of publications, see e.g. 6.3.9. The manuscripts include both results on the basic theory for such distance functions and "optimal'' (minimizing the rotational dependency) neighbourhood sequences, see Figure 2.
In a distance transform (DT), each picture element in an object is labeled with the distance to the closest element in the background. Thus the shape of the object is "structured" in a useful way. Only local operations are used, even if the results are global distances. DTs are very useful tools in many types of image analysis, from simple noise removal to advanced shape recognition. In the DT obtained when using distances based on neighbourhood sequences, the distance values consitutes a layer-by-layer structure. The set of grid points having the same distance value constitutes a Jordan surface (or several Jordan surfaces). A Jordan surface separates the the set of grid points into two connected sets
|
|
Céline Fouard, Magnus Gedda
Funding: SLU S Faculty; UU TN Faculty
Period: 0601-
Abstract: In several application projects we have discovered the benefit of computing distances weighted by the gray levels traversed, e.g., project 12. There are many ways of doing this, and in this project we have made a thorough comparison of the distances calculated with Gray Weighted Distance Transforms (GWDT) and the Weighted Distance Transforms On Curved Spaces (WDTOCS). A small example of shortest paths is found in Figure 3. The work was presented at Discrete Geometry for Computer Imagery (DGCI'06) and published in the proceedings from the conference. The next step is to do a through examination of the performance of the underlying algorithms in these calculations.
 |
Robin Strand, Gunilla Borgefors
Funding: Graduate School in Mathematics and Computing (FMB)
Period: 0308-
Partners: Christer Kiselman, Dept. of Mathematics, UU; Peer Stelldinger, University of Hamburg, Germany; Benedek Nagy, Dept. of Computer Science, Faculty of Informatics, University of Debrecen, Debrecen, Hungary
Abstract: The main goal of the project is to develop image analysis and processing methods for volume images digitized in non-standard 3D grids. Volume images are usually captured in one of two ways: either the object is sliced (mechanically or optically) and the slices put together into a volume or the image is computed from raw data, e.g., X-ray or magnetic tomography. In both cases, voxels are usually box-shaped, as the within slice resolution is higher than the between slice distance. An image acquisition method, the Direct Fourier method has been developed for non-standard grids during 2006, see 6.3.14.
Before applying image analysis algorithms, the images are usually interpolated into the cubic grid. However, the cubic grid might not be the best choice. In two dimensions, it has been demonstrated in many ways that the hexagonal grid is theoretically better than the square grid. The body-centered cubic (bcc) grid and the face-centered cubic (fcc) grid are the generalizations to 3D of the hexagonal grid. In the bcc grid, the voxels consist of truncated octahedra, and in the fcc grid, the voxels consist of rhombic dodecahedra. The fcc grid is a densest packing, meaning that the grid points are positioned in an optimally dense arrangement. The fcc and bcc grids are reciprocal, so the Fourier transform on an fcc grid results in a bcc grid. In some situations, the densest packing (fcc grid) is preferably in the frequency domain, resulting in a bcc grid in spatial domain. In some cases, the densest packing is prefered in the spatial domain.
Some results about topology preserving digitization with the fcc and bcc grids were presented at IWCIA (see 6.3.13). The results show that especially the fcc grid is by far better than the cubic grid in this aspect. Other aspects of topology-preserving digitization on the fcc grid have also been examined, but are not yet published.
Ingela Nyström, Joakim Lindblad, Gunilla Borgefors
Funding: SLU S Faculty, UU TN Faculty
Period: 0109-
Partners: Nataša Sladoje (Matic), Faculty of Engineering, University of Novi Sad, Serbia;
Abstract: The advantages of representing objects in images as fuzzy spatial sets are numerous and have lead to increased interest for fuzzy approaches in image analysis. Fuzziness is an intrinsic property of images and a natural outcome of most imaging devices. Preservation of fuzziness implies preservation of important information about objects and images. Our previous results within this project show that an improved precision of a shape description can be achieved if the description is based on a fuzzy, instead of a crisp shape representation, where the fuzzy membership of a point reflects the level to which that point belongs to the object.
During 2006, a manuscript on the representation and reconstruction of fuzzy disks by moments, containing derivation of theoretical error bounds for the accuracy of the estimation of moments of a continuous fuzzy disk from the moments of its digitization, as well as showing that, for a certain class of membership functions, there exists a one-to-one correspondence between the set of fuzzy disks and the set of their generalized moment representations, was accepted for publication in the Fuzzy Sets and Systems journal.
Ingela Nyström, Joakim Lindblad, Stina Svensson
Funding: SLU S Faculty, UU TN Faculty
Period: 0301-
Partners: Nataša Sladoje (Matic) and Tibor Lukic, Faculty of Engineering, University of Novi Sad, Serbia
Abstract: This project concerns the development of a method for feature based defuzzification of spatial fuzzy sets. The developed method generates crisp shapes from fuzzy shapes by finding a crisp shape at a minimal distance to the fuzzy shape. We define the distance between two fuzzy sets as a distance between their feature-based representations in a chosen feature space. We have found it appropriate for defuzzification to incorporate both local and global features of the two sets. We have studied the use of membership values, gradient, area, perimeter, and centre of gravity in the distance measure. Several existing distance measures can be used to define the distance measure in the feature space. We have so far focused the research on Minkowski type distances measure.
The defuzzification method was further developed during 2006. A method for generating the crisp discrete representation of a fuzzy set at an increased spatial resolution, compared to the resolution of the fuzzy set, was developed and presented at the IWCIA conference in June 2006. Additional refinement of the method was achieved by the use of a scale space approach, providing preservation of feature values over a range of scales in the defuzzification process. Initial results from such an approach, where area at a range of scales was used in the feature distance, were presented at the DGCI conference in October 2006. That presentation also included a practical implementation of the method for 3D data. An example of defuzzification of a 3D data set at increased resolution is presented in Figure 4.
Ongoing research regarding improvements of the optimization part of the method, as well as practical application of the defuzzification method to Cryo-ET data of proteins (see Project 12), was also undertaken during 2006.
|
|
Stina Svensson, Magnus Gedda
Funding: The Swedish Research Council (project 621-2005-5540); SLU S Faculty; UU TN Faculty
Period: 9801-
Abstract: Methods for decomposition of 3D discrete objects as well as grey-level representations of proteins (see Project 12) have earlier been developed at CBA. These methods have been further developed by utilising the concept of fuzzy sets. The application in mind is Cryo-ET data of proteins, but the method is general and can be used as a blob separation algorithm for 2D or 3D grey-level images in applications where grey-levels are increasing towards the internal parts of the blobs. By using fuzzy sets, the inner properties of the structure is enhanced, thus, aiding decomposition. The decomposition scheme combines fuzzy distance information from the fuzzy object and fuzzy distance based hierarchical clustering of local maxima (see Project 3) with a region growing process to identify the parts of the fuzzy object. This approach shows promising results. An article describing the theoretical part of this work was accepted to Pattern Recognition Letters and available on-line during 2006.
Stina Svensson
Funding: The Swedish Research Council (project 621-2005-5540); SLU S Faculty
Period: 0601-
Partner: Ida-Maria Sintorn, CSIRO Mathematical and Information Sciences, North Ryde (Sydney), Australia
Abstract: Chamfer matching is a template matching method based on geometric image features and can be used for both 2D and 3D images. It finds good fits between the template and edges in a search image. A generalized cost function between the edges in the search image and the template, a list of coordinate pairs corresponding to the searched pattern, is minimized. To guide the template to good positions, a distance transform (DT) is calculated from edges in the search image and the sum of the distance values hit by the superimposed template constitute the cost function. Translation, scaling, rotation, and perspective changes are for 2D images and translation, scaling, and rotation for 3D images. By embedding the chamfer matching in a resolution hierarchy (hierarchical chamfer matching algorithm, HCMA), the algorithm results in a fast, general and robust matching algorithm.
In this project, modifications of HCMA to even further improve its robustness are investigated. A first step is to use a distance weighted propagation of gradient magnitude (GM) as a cost image instead of the distance transform (DT) of a binarised edge image. The benefits are that no binarisation of the gradient magnitude image is needed, hence removing one step in the process and reducing the risk of loosing "true'' match positions by a poor binarisation method. This approach, hierarchical chamfer matching based on propagation of gradient strengths (GM-HCMA), was presented at Discrete Geometry for Computer Imagery (DGCI'06) and published in the proceedings from the conference. GM-HCMA also applied in Project 12, showing good results.
Bo Nordin, Ewert Bengtsson
Funding: UU TN Faculty
Period: 8807-
Abstract: In recognition of the need in image analysis research to have a good platform for interactive work with digital images, we several years ago started a project with the aim of developing such a platform. The project originally involved some 10 man years of work, which would have been impossible to finance by regular research money. But through a cooperation with a group of companies we co-ordinated our interests of obtaining a good software platform for research with their interest in development of a new software product. Unfortunately, the companies never actively turned the resulting system, which was given the name IMP, into a product. At CBA, however, the IMP system has been used as a software basis for most of the teaching and research in image analysis for the last decade.
Some years ago, we started a major revision of the system as a "background task'' for Nordin. The main goal was to re-program the core system in C++ to make it easier to maintain and extend. In 2002, we decided to write a completely new program platform, Pixy, based on the new C++ core and with all image analysis functions written in C++ in order to take advantage of the C++-specific language constructs (classes, inheritance, polymorphism, templates, etc.) to enhance the programmer's API and make the code more reusable.
In Pixy, it is easy to add plug-in modules with new functionality and new classes: several such modules have been implemented: MUSE (multivariate segmentation) and filter editors for editing filters in the spatial domain as well as in the Fourier domain. A first test version of Pixy was released internally at CBA during 2003 and a more complete version was released during 2006.
Analysis of microscopic biomedical images
Gunilla Borgefors, Joakim Lindblad, Hamid Sarve
Funding: SLU, S Faculty, Swedish Research Council
Period: 0503-
Partner: Carina Johansson, Dept. of Clinical Medicine, Örebro University
Abstract: In order to evaluate how tissue reacts on implants, the interface between the implant and the tissue must be studied. Today, this procedure is done manually in a microscope.
The aim of this project is to develop automatized image analysis methods for analyzing images of the junction of tissue and implant. This method shall make the procedure more effective as well as giving an objective estimation.
The analysis involves segmentation of the images in different tissue-types and measurement of some relevant measures such as length, area and volume.
Before the analysis, methods that shall remove artifacts be applied. Differences in graylevels, color and possibly texture features will be used for the recognition. Known methods will be used to present the result. The interpretation of the values however, will not be done by the postgraduate student.
This project will result in a number of publications (at conferences and in technical and medical journals) about the new methods used as well as the resulting measurement.
Patrick Karlsson, Ewert Bengtsson, Joakim Lindblad, Gunilla Borgefors
Period: 0603-
Partners: Anna-Stina Höglund, Jingxia Liu, Lars Larsson, Dept. of Neuroscience, UU
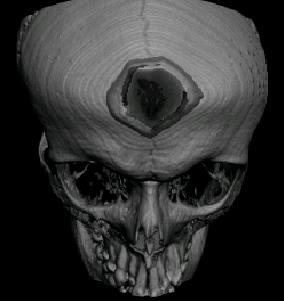
Abstract: The need for understanding of the three dimensional (3D) spatial arrangement of myonuclei in skeletal muscle fibers is great. A highly detailed 3D spatial description of the organization of myonuclei in healthy and diseased human muscle cells enables detailed understanding of the underlying mechanisms of muscle wasting associated with, e.g., neuromuscular disorders, and aging. The current poor understanding of the spatial arrangement of myonuclei is to be remedied by an interdisciplinary collaboration between the CBA and the Muscle Research Group (MRG) at UU. This project develops and evaluates methods for modeling and quantitative analysis of 3D distributions of myonuclei by utilizing the proficiency of modern confocal microscopic techniques to create true 3D volume images. Advanced computerized modeling of the elongated generalized cylinder structure of the imaged muscle cells is paramount in investigating the myonuclear domain, i.e., each finite volume in which a myonucleus control the gene products. The complex topological conditions put on 3D models and measurement methods, compared to 2D equivalents, are a challenge in developing appropriate image analysis tools and algorithms. The project was presented with a poster presentation at "Medicinteknikdagarna 2006'' in Uppsala.
|
Stina Svensson, Magnus Gedda
Funding: The Swedish Research Council (project 621-2005-5540); SLU S Faculty; UU TN Faculty
Period: 0401-
Partners: Dept. of Cell and Molecular Biology (CMB), Karolinska Institute, Stockholm; Dipartimento di Energetica "S. Stecco'' and Dipartimento di Fisica, Universitá di Firenze (UF), Florence, Italy; Laboratoire de Biophysique (LB), Statistique ITP/SB, Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland, Medical Research Council Centre (MRCC), Cambridge, United Kingdom.
Abstract: State of the art imaging techniques makes it possible to study individual proteins and other macromolecules from a structural point of view. Descriptions with respect to geometry and shape facilitates studying protein dynamics. This type of study is essential to increase the understanding of their biological role. CMB has developed methods, using cryo electron tomography, for 3D imaging of individual proteins at a resolution of approximately 2 nm. Moreover, they have together with UF and LB developed methods for modelling protein dynamic based on the images. Fitting a model to each protein in the image has so far been done manually. For large-scale studies, computerized image analysis serves as an essential tool to automatically and objectively fit the protein model to image content. In this project, we develop methods to fill the gap between image and model and thereby make large-scale studies of the movement of proteins possible. We will develop methods for automatically extracting the geometrical features needed as input to the models. This will be done taking into account both grey-level information (which reflects the internal structure of the protein) and 3D shape information.
The first step in this process is to identify significant parts of the protein. This is of interest as the parts and their relative position is the key to understanding how flexible a protein is and how it can interact with, or bind to, other proteins or substances. During 2006, a method, theoretically described in Project 7 was presented at The International Conference on Pattern Recognition (ICPR) and published in the proceedings from the conference. The method was applied the Immunoglobulin G (IgG) antibody imaged by Cryo-ET. The IgG antibody consists of three parts, the Fc stem and the two Fab arms, and has a volume of 1500 voxels in the given sampling. The parts are approximately equal in size. In some cases, the connection (hinge) between a Fab and the Fc stem is not visible due to the resolution. Example are shown in Figure 6.
The next step is to extract relevant geometrical features. This work was, for the IgG antibody, initiated during 2006 in cooperation with UF and LB. The features of interest are the interdomain angles and the translation of the domains with respet to each others.
Moreover, we have recently started to investigate another protein, namely the Met tyrosine kinase receptor. This is done in cooperation with with MRCC, UF, and LB. The Met protein controls growth, invasion, and metastasis in cancer cells and activating Met mutations predispose to human cancer. It has a unique biological role and is therefore of interest to study. In its mature form, Met consists of an extracellular ![]() chain and a longer
chain and a longer ![]() chain. The
chain. The ![]() chain and the first part of the
chain and the first part of the ![]() chain folds to a
chain folds to a ![]() propeller structure. This
propeller structure. This ![]() propeller region is sufficient for the binding to its ligand, hepatocyte growth factor / scatter factor (HGF/SF). The remaining part of the
propeller region is sufficient for the binding to its ligand, hepatocyte growth factor / scatter factor (HGF/SF). The remaining part of the ![]() chain is called stalk, due to its stalk-like structure. The stalk is, for the moment, understood to hold the
chain is called stalk, due to its stalk-like structure. The stalk is, for the moment, understood to hold the ![]() propeller in correct orientation for the ligand binding. To get a better understanding of the biology behind, a quantitative study of the flexibility of the stalk for Met. During 2006, we have, using an approach similar to the above described, developed a method for identifying the
propeller in correct orientation for the ligand binding. To get a better understanding of the biology behind, a quantitative study of the flexibility of the stalk for Met. During 2006, we have, using an approach similar to the above described, developed a method for identifying the ![]() propeller and the stalk. Furthermore, we have extracted a representation, a stalk curve suitable for extracting geometrical features regarding its flexibility. This work is described in a manuscript late 2006.
propeller and the stalk. Furthermore, we have extracted a representation, a stalk curve suitable for extracting geometrical features regarding its flexibility. This work is described in a manuscript late 2006.
To find structures of interest in Cryo-ET data, a matching algorithm can be used. During 2006, an algorithm -- Hierarchical chamfer matching based on propagation of gradient strengths (GM-HCMA) -- was developed, see Project 8, and tested on Cryo-ET data of the IgG antibody. A template created from the Protein Data Bank was used. The results are illustrated in Figure 7. The work was presented at Discrete Geometry for Computer Imagery (DGCI'06) and published in the proceedings from the conference.
Amalka Pinidiyaarachchi, Carolina Wählby, Ewert Bengtsson
Funding: SIDA
Period: 0605-
Partner: N. Rantatunga, Dept. of Pathology, University of Peradeniya, Sri Lanka
Abstract: In many biomedical applications quantitative measurements of stained nuclei of both cytological and histological samples are used in aid of decision making by the expert. The digital images produced in such processes are in color and segmentation of specific regions of such images is an area that has been widely studied. A project is carried out in collaboration with the Dept. of Pathology, Faculty of Medicine, University of Peradeniya, Sri Lanka where thyroid smear samples are expected be analyzed. The image processing goal to be achieved is the development of successful color image segmentation methods. This involves tests in different color spaces, selection of a good segmentation method and dealing with heavily clustered cell nuclei segmentation. The biological goal in the study is to use the methods effectively in extracting various features of cell nuclei that can be used in expert decision making process. Figures 8 shows two sample images and the intermediate segmentation results after initial segmentation using color information followed by seeded watershed transform. Extensions of watersheds to color images where color gradient measures and statistical comparison with neighborhood are used are to be tested on the images in further studies.
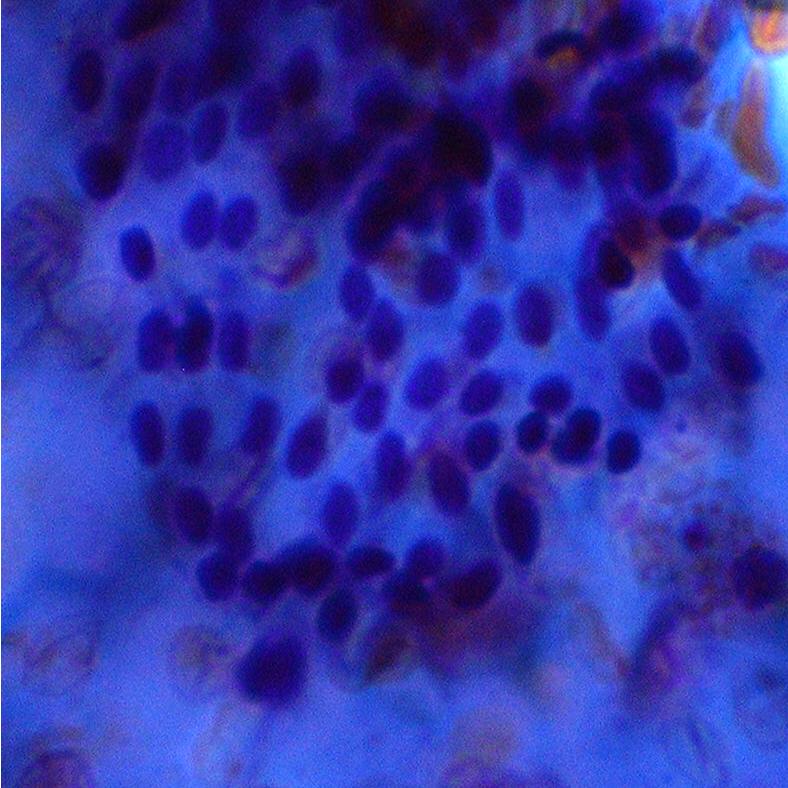
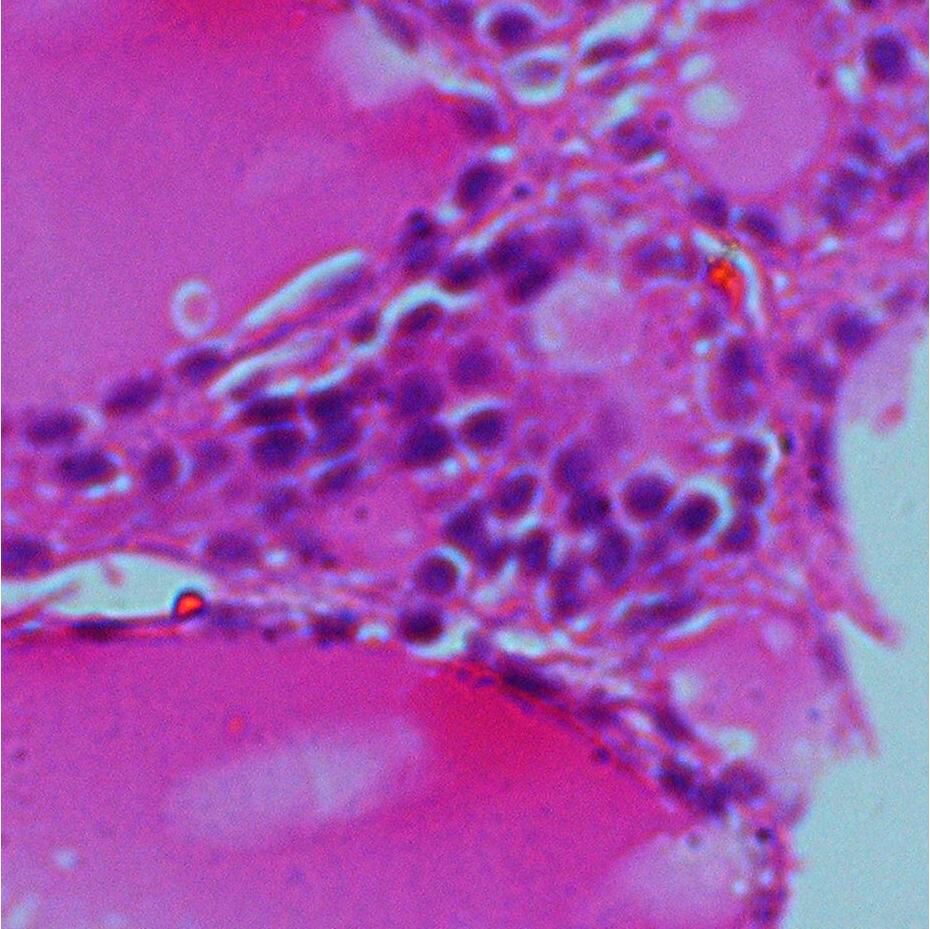

|
Amalka Pinidiyaarachchi, Patrick Karlsson, Carolina Wählby, Ewert Bengtsson
Funding: UU TN Faculty, SIDA
Period: 0305-
Partners: Malin Jarvius, Chatarina Larsson, Mats Nilsson, Ola Söderberg, and Irene Weibrecht, Dept. of Genetics and Pathology, UU
Abstract: The interior of a cell is elaborately subdivided into many functionally distinct compartments, often organized into intricate systems. One way of studying such compartments is by the use of different fluorescent markers that bind specifically to the objects of interest. This type of staining followed by imaging through a microscope often results in point-source signals, or "blobs", together with a background of noise and autofluorescence. 3D images are acquired by making non-invasive serial optical sections of the object. Analysis of spatial relationships in 2D and 3D requires pre-processing followed by separation and segmentation of the different blobs by combining intensity and shape information. Once the different blobs are detected, the goal is to detect spatial relationships and non-random patterns in the blob distribution.
Carolina Wählby, Amin Allalou
Period: 0608-
Funding: EU-Strep project ENLIGHT (ENhanced LIGase based Histochemical Techniques)
Partners: Anton K. Raap, Frans M. van de Rijke, Roos Jahangir Tafrechi, Dept. of Molecular Cell Biology, Leiden University Medical Center, The Netherlands; Visiopharm, Hørsholm, Denmark
Abstract: Cell cultures as well as cells in tissue always display a certain degree of variability, and measurements based on cell averages will miss important information contained in a heterogeneous population. An automated method for image based measurements of mitochondiral DNA (mtDNA) mutations in individual cells has been implemented. The mitochondria are present in the cell's cytoplasm, and each cytoplasm has to be delineated. Three different methods for segmentation of cytoplasms were compared and it was shown that automated cytoplasmic delineation can be performed 30 times faster than manual delineation, with an accuracy as high as 87%. The final image based measurements of mitochondrial mutation load were also compared to, and showed high agreement with, measurements made using biochemical techniques. The development of image based single cell analysis will continue to be in focus in the ongoing ENLIGHT project. See also Figure 9.
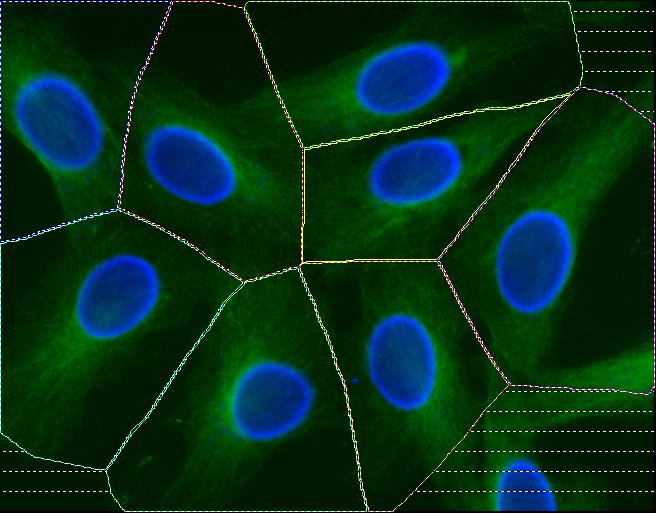
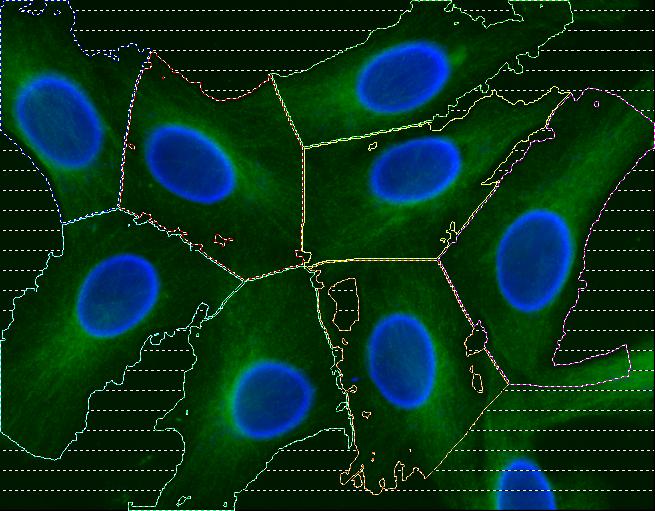
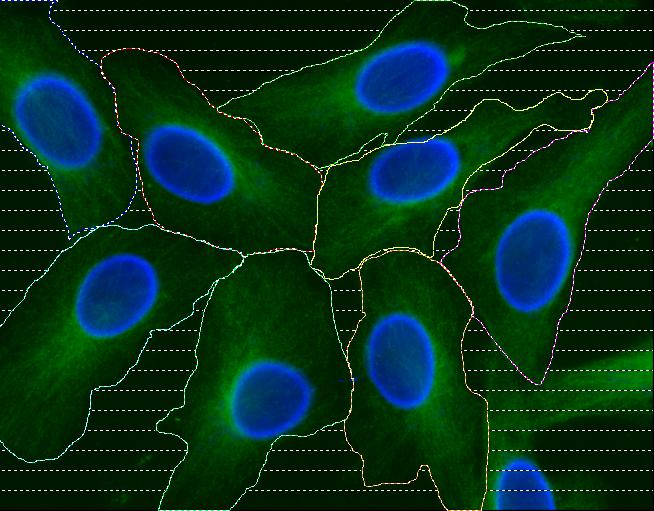
|
Milan Gavrilovic and Carolina Wählby
Period: 0611-
Funding: EU-Strep project ENLIGHT
Abstract: In fluorescence microscopy, during acquisition of multiply labeled specimen, two or more of the emission signals can often be physically located in the same area or very near to one another in the final image due to their close proximity within the microscopic structure. This effect is known as colocalization. Since red and green wavelengths are usually selected, if spectral overlap occurs, these two wavelengths will appear as yellow emission. Existing methods for quantification and localization of colocalized pixels have been implemented and compared. At the same time work has been initiated on development of new methods based on spectral decomposition of reference spectra.
Carolina Wählby
Funding: Uppsala BioX
Period: 0501-
Abstract: Nilsson et al. are developing new methodism for molecular analyses that allow analysis at the ultimate level of single bio-molecules through padlock- and proximity probing coupled to rolling-circle amplification. Rolling-circle products spontaneously form micron-sized coils (blobs) that can be immobilized randomly on a glass surface, referred to as a random blob array. Blobs are identified by hybridization of fluorescence labelled tag-oligonucleotides. Therefore, individual detection oligonucleotides act as biotransistors that convey and amplify the information from the nanometer-sized probe molecules to observable micron-sized products. In this project, the goal is to use combinations of fluorescent molecules on single detection oligonucleotides, as well as repeated staining and de-staining, in order to create image data that can be analysed for simultaneus identification of many different types of DNA fragments, transcripts, and proteins. The position of the reaction sites of a random blob array are random. In order to classify the blobs, they are first detected by a combination of filtering and morphological operations. Methods based on spectral decomposition are therafter used for blob classification.
3D analysis and visualization
Erik Vidholm, Ewert Bengtsson, Ingela Nyström, Stefan Seipel, Filip Malmberg
Funding: Swedish Research Council, UU TN Faculty
Period: 0301-
Partners: Lennart Thurfjell, GE Healthcare, Uppsala/London, UK; Gunnar Jansson, Dept. of Psychology, UU; Hans Frimmel, Dept. of Oncology, Radiology, and Clinical Immunology, UU
Abstract: Modern medical imaging techniques provide 3D images of increasing complexity. Better ways of exploring these images for diagnostic and treatment planning purposes are needed. Combined stereoscopic and haptic display of the images form a powerful platform for such image analysis.
In order to work with specific patient cases, it is necessary to be able to work directly with the medical image volume and to generate the relevant 3D structures directly as they are needed for the visualization. Most work so far on haptic display has used predefined object surface models. In this project, we are creating the tools necessary for effective interactive exploration of complex medical image volumes for diagnostic or treatment planning purposes through combined use of haptic and 3D stereoscopic display techniques. The developed methods are tested on real medical application data.
Our current applications are interactive liver segmentation from CT images, see Project 19, accelerating the computation of 3D gradient vector flow fields, see Project 22, and hardware assisted visualization of breast MR images, see Project 20. In additon to this, we are working on haptic interaction with 3D deformable surface meshes and 3D Live wire from a more theoretical point of view.

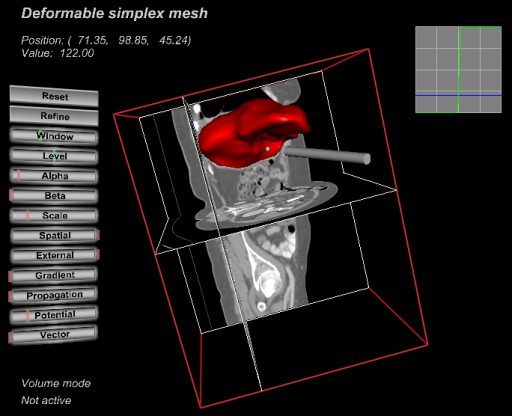
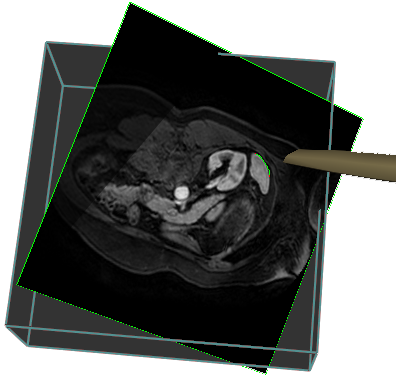
|
Erik Vidholm, Ingela Nyström, Ewert Bengtsson
Funding: Swedish Research Council, UU TN Faculty
Period: 0501-
Partners: Sven Nilsson, Hans Frimmel, Dept. of Oncology, Radiology, and Clinical Immunology, UU
Abstract: The manual step in semi-automatic segmentation of medical volume images typically involves initialization procedures, such as placement of seed-points or positioning of surface models inside the object to be segmented. The initialization is then used as input to an automatic segmentation algorithm. We investigate how such initialization tasks can be facilitated by using haptic feedback.
In this project, we develop interactive methods for segmenting the liver from CT scans of patients with neuroendocrine tumors. Liver segmentation is of importance in hepatic surgery planning, where it is a first step in the process of finding vessels and tumours, and the classification of liver segments. Liver segmentation may also be useful for monitoring patients with liver metastases, where disease progress is correlated to enlargement of the liver.
We have used the fast marching algorithm, where haptics is used to make the initialization of the algorithm easier and more efficient. Two users placed initializations in 52 datasets (26 patients at two different occasions) using the haptic user interface. The mean interaction time is about 45 seconds per dataset and the resulting segmentations highly correlate between the users, i.e., the method has high precision. The results from this project was presented at Medical Image Computing and Computer Assisted Intervention (MICCAI'06) in October. The next step is to verify the accuracy of the method.
Ewert Bengtsson, Erik Vidholm, Ingela Nyström
Funding: UU TN Faculty, The Australian Research Council
Period: 0503-
Partners: Stuart Crozier, Andrew Mehnert and co-workers at ITEE department, University of Queensland, Brisbane, Australia; Ivo Hanak, Dept. of Computer Science, University of West Bohemia, Czech Republic
Abstract: The pattern of change of signal intensity over time in contrast enhanced magnetic resonance (MR) images of the breast is a useful indicator of malignancy. The methods used for assesing and visualizing this in current clinical practice are rather tedious; it is difficult to visualise and evaluate 4D (3D volumes over time) data effectively. In this project, we are developing and evaluating improved methods for such visualization and evaluation. The project started during Bengtsson's sabbatical at the University of Queensland in 2004-2005 and continued as a joint project after his return to CBA. In September 2006, Erik Vidholm was visiting Andrew Mehnert at the department in Brisbane.
During 2006, we have developed a program where the 4D dataset is visualized with hardware accelerated maximum intensity projection (MIP) in the hue-saturation-value (HSV) colour space. The program also allows for drawing regions of interest.
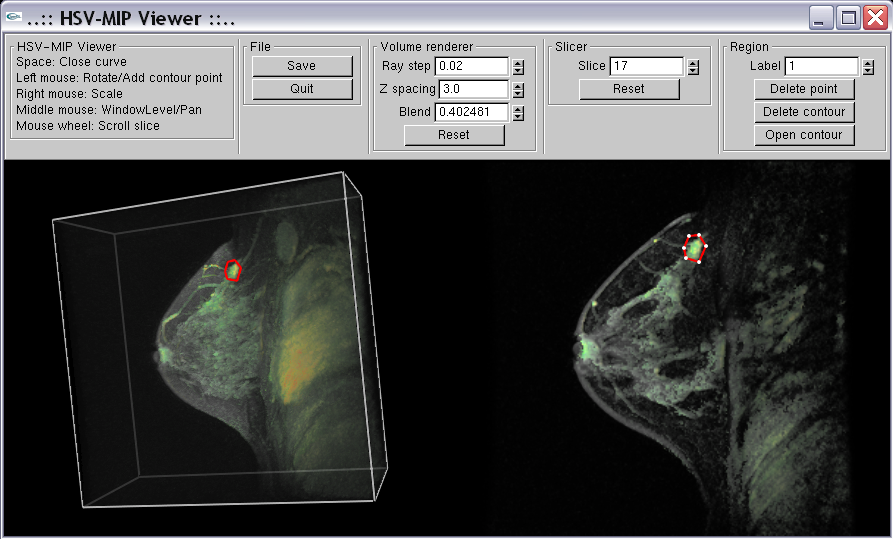
|
Suthakar Somaskandan, Ingela Nyström, Ewert Bengtsson
Funding: SIDA
Period: 0409-
Abstract: Several of the modern imaging systems provide 3D volume information, e.g., CT, MRT, SPECT, and ultrasound. This is very useful since the human body is 3D. However, to reach a diagnostic conclusion, the 3D images need to be projected onto the 2D computer screens in more sophisticated ways than slice by slice projections.
It is particularly useful to device methods where the user interactively can explore the 3D information in the images. Earlier, allowing dynamic interaction with medical volume images of realistic, clinically useful resolution required very expensive display stations driven by high-performance computers. Today, a PC equipped with a high-end standard graphics card (mass produced for the game market) can be used quite effectively for the purpose of medical visualization. Still much research work is needed to find out the most effective way of using such display facilities for exploring medical data. The research task in this project is to develop such display methods, e.g., by utilizing the programmability of today's graphics cards.
Erik Vidholm, Ingela Nyström, Ewert Bengtsson
Funding: Swedish Research Council, UU TN Faculty
Period: 0508-
Partner: Per Sundqvist, Dept. of Information Technology, UU
Abstract: In our work on interactive 3D segmentation, see Project 18, we have developed methods to facilitate initialization of our segmentation algorithms. One way is to base the haptic interaction on gradient vector flow (GVF) that propagates edge information from strong boundaries into the center of objects. This approach allows a user to feel object boundaries while still being centered inside the object. The GVF field can also be used to drive a deformable model, and then we get an intuitive connection between the model and the interaction.
The computation of a GVF field consists mainly of solving a huge discretized system of elliptical partial differential equations (PDEs). The convergence rate of the commonly used numerical scheme to compute GVF does not allow for practical use in 3D applications. This project aims at presenting alternative computation schemes to speed up the computation. The discrete equations have several properties that can be utilized to accelerate the process of finding an approximate solution. We investigate how stationary iterative methods, preconditioned conjugate gradient methods, and multigrid methods can be tuned to solve our problem.
Our results show that it is possible to obtain much better performance by only small modifications of the original scheme. The results also show that the multigrid algorithm is the fastest and allows us to compute the GVF field in the order of one minute for a standard medical image on a standard computer (compared to approximately one hour with the commonly used approach). The results were presented at the International Conference on Pattern Recognition (ICPR'06) in August.
Ewert Bengtsson
Funding: UU TN Faculty
Period: 9911-
Partners: Anders Hast, Dept. of Mathematics, Natural Sciences, and Computing, University College of Gävle; Tony Barrera, Barrera Kristiansen AB, Uppsala
Abstract: Computer graphics is increasingly being used to create ealistic images of 3D objects. Typical applications are in entertainment (animated films, games), commerce (showing 3D images of products on the web which can be manipulated and rotated), industrial design, and medicine. For the images to look realistic high quality shading and surface texture and topology rendering is necessary. Many fundamental algorithms in this field were developed already in the early seventies. The algorithms that produce the best results are computationally quite demanding (e.g., Phong shading) while other produce less satisfactory results (e.g., Gouraud shading). In order to make full 3D animation on standard computers feasible, high efficiency is necessary. We are in this project re-examining those algorithms and are finding new mathematical ways of simplifying the expressions and increasing the implementation speeds without sacrificing image quality.
The project is carried out in close collaboration with Tony Barrera. It has been running since 1999 and resulted in 2004 in a PhD thesis by Anders Hast. By that time it had produced 20 international publications. Since then another book chapter and a few reviewed conference papers have been produced.
Stefan Seipel
Funding: UU TN Faculty
Period: 0603-0608
Comment: Anders Nivfors (MSc student) is a cooperation partner.
Abstract: Ice is a common phenomenon in nature and occurs in our everyday surroundings in form of e.g. icicles and ice cubes. The rendering of ice is, however, a field of real-time computer graphics that has not been subject to exhaustive research. Realistic rendering of natural phenomena in real-time has always been one of the most difficult tasks. As a result of this there are numerous papers describing techniques to implement fire, smoke, clouds, fog, water etc, but ice seems to be all ignored. In this project we identify the most eye catching characteristics of ice and what distinguishes ice from similar materials such as glass. The main focus of the project is to develop a method for rendering ice and its most important characteristics in real-time using the functionalities of the GPU on modern graphics cards. The methods developed are capable of filling a given convex geometry with air particles and bubbles as well as adding an arbitrary amount of cracks. They comprise the creation of a bumpy and irregular surface that reflects and refracts the environment. An improved image space technique for clipping a geometry using the Boolean difference of two geometries was developed, as well. It is applied to the cracks in order to clip them against the ices geometry. Reflection and refraction effects on the ice were realized by using environment mapping. An improved method for two-sided refraction was developed that combines the normals of the front and back side of the ice object. See Figure 12.

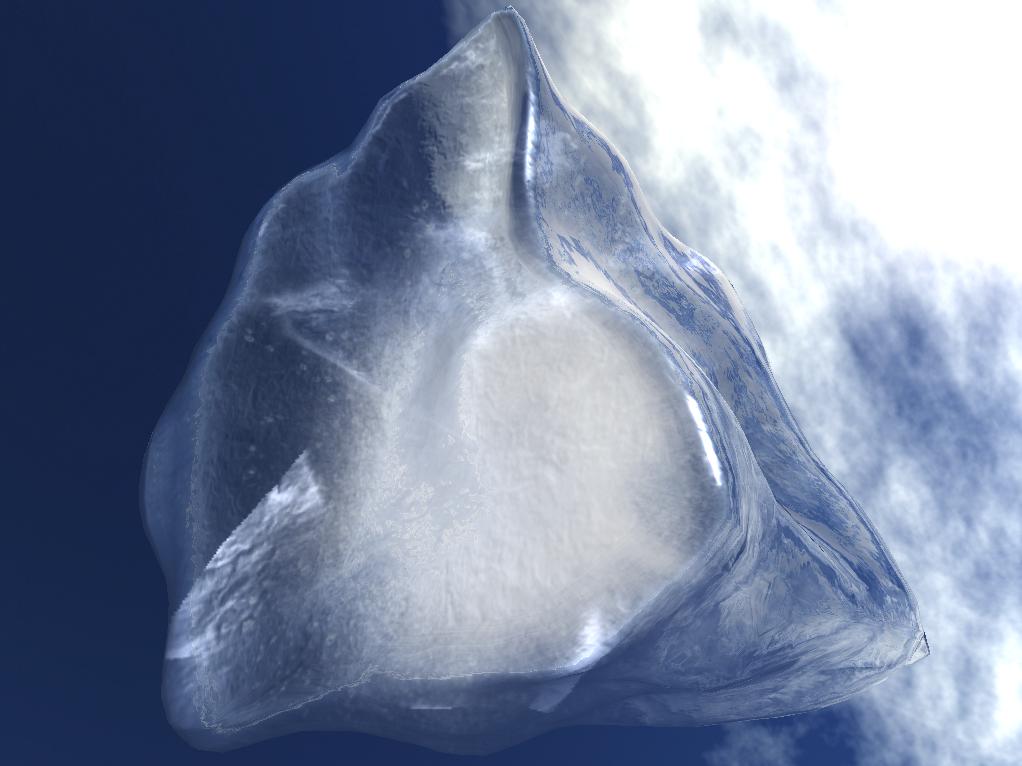
|
Stefan Seipel
Funding: UU TN Faculty
Period: 0606-0612
Comment: Cezary Bloch (MSc student) is a cooperation partner.
Abstract: In this project we aimed at developing new methods to directly render multivariate volume data. When rendering 3D scalar fields (e.g. in medicine) on a 2D computer screen, the means for expressing various attributes in every voxel are usually limited to colors, only. Graphically more complex annotations or glyphs require a too large footprint on-screen and hamper perception of the spatial structure of the underlying data. In this project we looked at new methods direct visualization of more than one scalar value, excluding color manipulations. The general objective was to investigate if a secondary variable could be visualized by modulating the illumination functions in 3D rendering, or by using modulated textures to express the secondary attribute. As a result of this experimental project, we came up with a new method that utilizes 3D normal mapping. A 3D normal map is used in the 3D shading model of the volume renderer to create volumetric bumps. The appearance of bumps is modulated with respect to the values of the additional scalar attribute. See Figure 13.
Stefan Seipel, Lars W. Pettersson
Funding: UU TN Faculty
Period: 0309-0612
Partners: Swedish Defense College; Dept. of Information Technology, UU; University College of Gävle
Abstract: In many important decision situations, more than only one stakeholder is concerned with the analysis and interpretation of the decisive data. Often, the data collected is very complex, it may be organized in several orthogonal dimensions or is ordered in multiple independent layers. Our technical means to efficiently visualize this complexity are limited to the two-dimensional grid of colored pixels on a computer screen. In this current project we investigate how we optimally can design visualizations that support more collaborating users who interact simultaneously with the same visualization in the same place. We use a rather unique display technology (in collaboration with and at the Swedish Defense College - FHS) to present up to 8 individual pictures at the same display surface. It allows to selectively controlling, which picture(s) are seen by individual users. In this environment we study the effects of several features of advanced visualizations, which are dynamic observer conditions, stereoscopic cues, and layer separated visualizations. See Figure 14.
|


|
Forestry related applications
Maria Axelsson, Filip Malmberg, Stina Svensson, Joakim Lindblad, Gunilla Borgefors
Funding: SLU S Faculty
Period: 0406-
Partners: Norwegian Pulp and Paper Research Institute (PFI), Trondheim, Norway; STFI-Packforsk, Stockholm; KTH Solid Mechanics, Stockholm; StoraEnso, Falun
Abstract: The internal structure of paper is important to study since many material properties correspond directly to the properties of single fibres and their interaction in the fibre network. How single fibres in paper bond and how this effects paper quality is not fully understood since most structure analysis of paper has been performed in cross sectional two dimensional (2D) images and paper is a complex three dimensional (3D) structure. Another application for wood-fibres that has recently gained a lot of interest is wood polymer composite materials. The properties of these materials do not only depend on the structure of the fibre network, but also on interaction between the fibres and the polymer matrix surrounding the fibres. Advances in imaging technology has made it possible to acquire 3D images of paper and wood polymer composite materials. In this project, image analysis methods for characterising the 3D material structure in such images are developed. The detailed knowledge of the material structure attainable with these methods is useful for improving material properties and for developing new materials.
An example slice from a binarised volume and a surface rendering of a sample of a composite material image with X-ray microtomography are shown in Figure 15.
The project objective is to achieve a complete segmentation of individual fibres and pores in volume images of the material. Given such a segmentation, any measurement of the internal structure is available. Measurements on individual fibres and the structural arrangement of fibres can then be related to macroscopical material properties. Other methods for measuring properties of the material, that do not require a complete segmentation of the samples, are also investigated.
In this project, different volume images of paper and composite materials are available for the studies. This includes one volume created from a series of 2D scanning electron microscopy (SEM) images at StoraEnso in Falun and X-ray microtomography volume images of paper and composite samples imaged at the European Radiation Synchrotron Facility (ESRF) in Grenoble, France. Furthermore, methods for creating other sample volume images are investigated.
During 2006, the project has resulted in a number of publications. A method for reducing ring artifacts in the volume images using orientation estimates and normalised convolution in the polar domain was presented at the DAGM conference in September 2006 and published in the Lecture Notes in Computer Science Series. See Figure 16 for an illustration of the method. At the Progress in Paper Physics Seminar three extended abstracts were presented. One abstract on 3D structure characterisation of newsprints, one on stress transfer and failure in pulp fibre reinforced composite materials and one on damage mechanisms in paper.
|
|
Maria Axelsson, Stina Svensson, Gunilla Borgefors
Funding: SLU S Faculty
Period: 0501-
Partners: STFI-Packforsk, Stockholm
Abstract: Press felts are used in the manufacturing of paper to press water from the wet fibre web after the sheet formation in a number of press nips. The press felt surface is a non uniform porous structure. The amount of water that can be pressed out of the fibre web and the amount of the water that is transfered back to the fibre web in the separation rewetting depends on the structure of the press felt surface.
In this project the amount of water that can be held in the interface pores between the press felt surface and the fibre web is estimated. Samples of press felts were compressed to different degrees against a glass plate and imaged in a Confocal Laser Scanning Microscope. A method for calculating the size of the interface pores by estimating the fibre web depression into the felt structure was developed. The method was published in Nordic Pulp and Paper Research Journal in 2006. And the method was also presented at SSBA 2006. For an example of 3D renderings of a press felt surface under load, see Figure 17.
|
|
|
|
Kristin Norell, Stina Svensson, Gunilla Borgefors
Funding: The Swedish Timber Measurement Council (VMR), SLU S Faculty
Period: 0505-
Partners: The Swedish Timber Measurement Council (VMR), Dept. of Forest Products and Markets, SLU
Abstract: The wood quality of a log can be analyzed to some extent by examining the log end. Such analysis is mostly performed manually at saw mills, where the log scaler has a couple of seconds to determine features like the approximate annual ring density, presence of rot and precense of compression wood. By using an image analysis application instead, the analysis can be more robust. In this project methods to measure important properties of logs in saw mill environment using computerized image analysis is developed. Some interesting features are:
- position of the center of the annual rings (pith)
- shape of the log end
- annual ring density
- rot
- blue stain
Pith position is found using filters to detect local orientation, Hough transform, and a final adjustment technique. Once the pith is found some other measurements will be easy, and others will be facilitated. Figure 18 shows two typical log end images. The result of the pith detection and the ground truth are marked with a white and a black +-sign, respectively. During 2006 the pith detection method has been presented at the Swedish Society for Automated Image Analysis Symposium and also submitted to a journal. Work has also been started on the detection of annual rings.


|
Kristin Norell, Stina Svensson
Funding: SLU S Faculty
Period: 0612-
Partners: Kim Dralle, Anders Björholm Dahl at Dralle A/S Cognitive Systems, Copenhagen, Denmark
Abstract: This project focuses on image analysis methods for identifying rot in log end faces. The purpose is to detect rot already while harvesting, or when the logs are in a stack waiting for transport. Logs are depicted using a standard color digital camera that can be mounted on a harvester or a vehicle. The goal is to find a robust method for detecting rot in timber suitable for practical use.
Joakim Lindblad, Gunilla Borgefors
Funding: The Swedish Farmers' Foundation for Agricultural Research (SLF)
Period: 0509-0603
Partners: AnalyCen Nordic AB, Lidköping; SeedGard AB (prev. Acanova), Uppsala; Anders Larsolle, Dept. of Biometry and Engineering, SLU, Uppsala
Abstract: ThermoSeed cereal seed treatment is a new method for thermal seed treatment developed by SeedGard AB (prev. Acanova). The method makes it possible to produce seed free from seed-borne pathogens without using chemical seed dressing. By exposing seeds for precisely conditioned hot humid air, pathogens are rendered harmless without affecting seed germinability.
It is of interest to facilitate objective and accurate monitoring of how different treatments and different types of stress affects the vitality of seeds. During the autumn of 2005 computer software for automatic segmentation and separation of individual plants as well as to measure relevant parameters, such as area and length, of the plants were developed. In early 2006, the software was delivered to SeedGard AB, where further testing and tuning of parameters followed.
Remote sensing
Tommy Lindell
Funding: UU TN Faculty, Dept. of Infrastructure, KTH, Stockholm
Period: 0406-
Partners: Kai Sörensen, Norwegian Institute for Water Research (NIVA); Terry Callaghan, Abisko Scientific Research Station (ANS), Fredrik Bergholm
Abstract: Field measurements were performed on Lake Torneträsk, and near Abisko. From June 2004, spectrometer measurements were run by Fredrik Bergholm on the arctic vegetation around Abisko Naturvetenskapliga Station (ANS), and spectral measurements were performed by Tommy Lindell and Kai Sörensen on Lake Torneträsk. The field measurements have continued during the following years 2005 and 2006. Fredrik Bergholm has been measuring with spectrometers, among other things from the ski lift in Abisko. Tommy Lindell and Kai Sörensen has been taking water samples for analyses, made manual recording of spectral characteristics and have had a floating device on Torne Träsk for spectral measurements during the summer months. Tommy Lindell has collected and analysed satellite data from the lake. Kai Sörensen has been using ships on Hurigrutten for continuous sampling of spectral information and water quality variables. These field measurements have contributed to the data base for the Scandinavian NorSEN project, which formally started in 2005. The project deals with validation of satellite images in polar-near areas, and build-up of ground- based spectral sensors around ANS. Little scientific work has been done before on the use of satellite data in polar-near areas, with low sun angles and special atmospheric conditions. The project aims to find normalisation and correction measures of the satellite data for these areas.
Tommy Lindell
Period: 0001-
Partners: CNR, Milan, Italy
Abstract: Test of the usefulness of air-borne digital camera and video for mapping water variables. Lindell has been constructing a holder for the digital video/camera for small aircrafts. Data have been collected from Lakes Erken and Mälaren, and from coral bottoms in Bisceyne National Park. Recently, tests of the usefulness of those images have been performed for the classification of the Swedish coastline. The system has been further used for the classification of the Swedish coastline.