|
At CDAC, a program for data collection and a screening program including image segmentation, artifact rejection, feature extraction and cell and specimen classification have been developed. The system has been used to collect and analyze data for well over 1000 cases so far and has reached an acceptable performance level as specified in the original project plan although based on analyzing less than 10% of the specimen area and needing about 10 times longer than acceptable for a final system to do that.
At CBA we initially focused on studying whether the 3D chromatin texture of the cervical cells can be utilized as a robust feature for detecting (pre-) cancerous lesions. For that purpose we scanned the cells at 40 different focus levels creating stacks of data for each nucleus. Dr. Sujathan a cytologist from RCC spent two periods each of two weeks in Uppsala and collected ground-truth annotated data.
In collaboration with Andrew Mehnert at MedTech West, Chalmers new texture analysis approaches were implemented and evaluated. We have also implemented several other feature extraction methods and adapted them for our data stacks. The conclusion is that the high quality perfectly focused images we can obtain through the stacks gives strong textural features for classification, but we have not been able to show any convincing advantages of using 3D data directly. We have also developed methods for evaluating low resolution images to find regions of high interest for a high resolution analysis and methods for rejecting artifacts initially segmented as nuclei. The results have been published as three different conference papers and a paper accepted for journal publication.
The Indian project is currently awaiting final evaluation. We are proposing a follow-up project where some of our improved texture analysis methods will be added to the Indian system at the same time as the hardware solution is redesigned to increase throughput with more than an order of magnitude. During 2012 we visited India for 9 days in April and held a workshop going through all the different parts of our systems and research results. Another visit and workshop is planned for early February 2013.
Martin Simonsson, Carolina Wählby
Partners: Johen Kreuger, Sara Thorslund, Gradientech AB, Uppsala
Funding: SciLife Lab Uppsala; eSSENCE; Dept. of IT, UU
Period: 1108-
Abstract: Tracking of cell movements in various cell culture setups is essential to many researchers in the life science sector. Gradientech AB, a Swedish biotech company, has developed CellDirector, a unique microfluidic system that academic researchers can use to study how concentration gradients of soluble proteins impact cell migration. The current project is focused on developing software for analyzing cell behavior and cell migration. The free open-source software CellProfiler developed at the Broad Institute will be used as a platform for a high-throughput system with automated high quality imaging, adapted for unlabeled cells, which are analyzed with regard to directionality of migration, speed, and acceleration. Apart from analyzing cell migration, the cell tracking aims at producing lineages, where cellular events such as cell division and cell death can be scored for single cells. A graphical user interface for visualizing and editing tracks imported from CellProfiler has been developed. This will be used for manual feed back in an iterative parameter optimization process, which aims to improve the automatic tracking. The progress of the project was presented in the poster session at BioImage Informatics 2012 in Dresden.
Carolina Wählby
Partners: Jeremie Zoueu, Olivier Bagui, Dept. Genie Electrique et Electronique, Institut National Polytechnique, Felix Houbhouet-Boigny, Cote d' Ivoire
Period: 1109-1201
Abstract: This project aims to propose an effective optical device based on LED spectral microscopy, which will be low cost, fast and easy to use in
the diagnosis of human malaria parasites, especially because the
sample will not need any special preparation or staining and the data
will be automatically processed to provide real-time diagnosis of the
type of the parasite, the parasitic density and its age for an
effective prescription. The collaborative project was initiated by a
3-month visit by Olivier Bagui, where we focused on the development of
efficient segmentation methods for unstained images of blood cells.
Andreas Kårsnäs, Martin Simonsson, Carolina Wählby, Robin Strand
Partners: Caroline Kampf, The Human Protein Atlas (HPA); Virginie Uhlmann, Imaging Platform, Broad Institute of Harvard and MIT, Cambridge, Massachusetts MA, USA; S. Issac Niwas, P. Palanisamy, Dept. of ECE, National Institute of Technology (NIT), Tiruchirappalli, India
Funding: SciLife Lab Uppsala
Period: 1201-
Abstract: The Human Protein Atlas (HPA) is an effort to map the location of all human proteins (http://www.proteinatlas.org/) and contains a large number of histological images of sections from human tissue. Methods for quantification of staining patterns in histopathology have many applications, ranging from antibody quality control to tumor grading. In this project we have tested a new method based on complex wavelets textural features as well as an approach inspired by WNDCHARM (Weighted Neighbor Distances using a Compound Hierarchy of Algorithms Representing Morphology) for classifying nuclear versus cytoplasmic staining. This project was presented at MICCAI 2012 workshop on Histopathology Image Analysis (HIMA): Image Computing in Digital Pathology, Nice, October 2012 and has also been accepted for publication in Journal of Pathology Informatics.
Martin Simonsson, Carolina Wählby
Partners: Anne Wuttke, Dept. of Medical Cell Biology, Uppsala University
Funding: SciLife Lab Uppsala
Period: 1211-
Abstract: Insulin secreting cells perform exocytosis and this can be detected with a GFP-modified protein as an increase in fluorescence signal. Time lapse sequences are acquired with a time interval of one second during one hour, observing changes in fluorescence signaling at different treatments of the cells. This results in huge data sets with more than 3000 images for a single experiment. The focus of this project is to extract relevant information from the image data and in an efficient way analyse and visualize the data.
Alexandra Pacureanu, Omer Ishaq, Carolina Wählby
Partners: Amin Allalou, Izolde AB, Uppsala; Johan Ledin, Evolutionary Biology Centre, Zebrafish platform, SciLife Lab Uppsala; Carlos Pardo, Mehmet F. Yanik, Research Laboratory of Electronics, Massachusetts Institute of Technology, Cambridge, USA
Funding: SciLife Lab Uppsala; TN-faculty, UU
Period: 1009-
Abstract: Isotropic 3D imaging of biological specimens is instrumental for further breakthroughs in life sciences. Many biological specimens with high relevance for basic research, disease studies and drug discovery, such as model organisms or 3D cell cultures, are semi-transparent to visible light. This lead to the advent of the technique dubbed optical projection tomography (OPT). The 3D internal structure is revealed by the attenuation variations of the light traversing the specimen. In OPT transverse slices of the specimen are reconstructed from a set of angular projections and stacked together into a volumetric image. This method enables in-vivo imaging of relatively large samples with high spatial resolution.
A high-throughput platform for cellular resolution, in vivo OPT of zebrafish has been developed at MIT, Cambridge, USA. With this system we have shown that OPT of zebrafish embryos can provide 3D information enabling high-throughput screening of subtle phenotypic changes in relation to drug treatment. However, OPT imaging systems in general are still quite sophisticated and costly. We are therefore developing a system for optical 3D isotropic imaging at microscopic scale, based on readily accessible hardware. The total price of the setup is kept under 1000 euros and the components can be easily obtained around the world. We have assembled the image acquisition system and acquired and reconstructed our first 3D images of zebrafish embryos. We are complementing the simple hardware with open source computational tools, embedding algorithms for image alignment, correction and reconstruction. Additionally, we are exploring the potential for compressed-sensing based volumetric reconstruction for optical projection tomography. Potential benefits include low-sensitivity to noise as well as a smaller number of required projections.
Our goal is to enable every life sciences research laboratory to have access to valuable 3D information on biological specimens.
Amin Allalou, Carolina Wählby
Partners: Nils-Göran Larsson, Dept. of Laboratory Medicine, Karolinska Institute; Mats Nilsson, Chatarina Larsson, Dept. of Genetics and Pathology, UU; Anton K. Raap, Roshan S. Jahangir Tafrechi, Frans M. van de Rijke, Karoly Szuhai, Raimond B. G. Ravelli, George M. C. Janssen, Dept. of Molecular Cell Biology, Leiden University Medical Centre, Leiden, The Netherlands; Angela Pyle, Patrick F. Chinnery, Wellcome Trust Centre for Mitochondrial Research, Newcastle University, Newcastle upon Tyne, United Kingdom; René F. M. de Coo, Dept. of Neurology, Erasmus Medical Center, Rotterdam, The Netherlands; Harsha K. Rajasimha, Center for Human Genetics Research, Vanderbilt University Medical Center, Nashville, Tennessee.
Funding: The Swedish Research Council, Collaboration Grant, Medicine
Period: 0801-
Abstract: Mutations of mitochondrial DNA (mtDNA) cause genetic syndromes with widely varying phenotypes and are also implicated in many age-associated diseases and the ageing process itself. Our knowledge of the principles governing segregation of mtDNA mutations in somatic tissues and in the germ line is very limited. In this collaborative project we combine a powerful technique for detection of individual mtDNA molecules with image analysis. We work with a variety of mouse models and the goal is to develop image analysis software to do three-dimensional (3D) reconstruction of the distribution of mutated mtDNA molecules in mammalian tissues. We want to use this technology to study segregation of mtDNA mutations in mouse tissues and to study the mtDNA bottleneck by visualizing the distribution of mutated mtDNA during oogenesis. The ultimate goal is to study the distribution of mtDNA mutations in embryos and placenta to establish principles for prenatal diagnosis. A paper describing the non-random segregation pattern was published in PlosOne.
Omer Ishaq, Alexandra Pacureanu, Carolina Wählby
Partners: Johan Elf, Gustaf Ullman, Dept. of Cell & Molecular Biology, Uppsala University
Funding: SciLife Lab Uppsala
Period: 1211-
Abstract: Stochastic optical reconstruction microscopy (STORM) is a super-resolution microscopy image acquisition technique for single-molecule localization. Like other stochastic super-resolution microscopy techniques it incorporates a trade-off between spatial- and temporal-resolution. Recently, a compressed-sensing (CS) based variant of STORM, called FasterSTORM, has been developed which substantially increases the temporal sampling of a stack of STORM image frames. This improvement is realized by increasing the density of activated fluorophores in each frame, followed by a subsequent CS-based retrieval of single-molecule positions even with overlapping fluorescent signals. However, the CS-based retrieval/decoding step is time consuming and can take as much as three hours for each image frame. We are working on accelerating the FasterSTORM method either through processing on a grid or by porting and processing it on the graphical processing units (GPUs). Additionally, we have tested and tried a number of L1-solvers for CS-based recovery of molecule positions.
Carolina Wählby, Alexandra Pacureanu, Marine Astruc
Partners: Mats Nilsson, Rongqin Ke, Marco Mignardi, SciLife Lab Stockholm
Funding: SciLife Lab Uppsala; TN-faculty, UU
Period: 1109-
Abstract: Typically, isolated individual cells are profiled on RNA level using single cell capture techniques, but these techniques are laborious and limited in terms of number of cells analyzed, and information about the histological context is lost. Our collaborators have developed a new molecular method, enabling in situ sequencing of mRNA, so that protein expression can be observed directly in tissue samples. We are developing image analysis tools for automated RNA sequencing, mapping, and visualization of gene expression patterns in tissue sections and investigated methods and software tools for unsupervised clustering (Figure 11).

|
Omer Ishaq, Alexandra Pacureanu, Carolina Wählby
Partners: Joseph Negri, Mark-Anthony Bray, Randall T. Peterson, Broad Institute of Harvard and MIT
Funding: SciLife Lab Uppsala
Period: 1203-
Abstract: Zebrafish (Danio rerio) is an important model organism in biomedical research due to its ease of handling and translucent body and consequently many human disease models have been established in the Zebrafish. Zebrafish embryos undergo spinal deformation upon exposure to chemical agents, such as Camptothecin (Cpt), that inhibit DNA repair. We are developing automated image-based quantification of spine deformation enabling whole-organism based assays for use in early-phase drug discovery campaigns. Our automated method for accurate high-throughput measurement of tail deformations in multi-fish micro-plate wells generates refined medial representations of partial tail-segments. Subsequently, these disjoint segments are analyzed and fused to generate complete Zebrafish tails. Based on these estimated tail curvatures we reach a classification accuracy of 91% on individual animals as compared to known control treatment. This accuracy is increased to 95% when combining scores for fish in the same well.
Alexandra Pacureanu, Carolina Wählby
Partners: Lena Claesson-Welsh, Xiujuan Li, Makoto Hayashi, Jeremy Adler, Eric Morin, Dept. of Immunology, Genetics and Pathology, Rudbeck Laboratory, SciLife Lab Uppsala
Funding: SciLife Lab Uppsala
Period: 1202-
Abstract: The formation of new blood vessels, angiogenesis, represents a key process in the growth of tissues from embryonic development to wound healing and cancerous tumors. Our collaborators are working on understanding how angiogenesis is induced, but also how it can be suppressed. We developed image analysis pipelines for several projects concerning assessment of endothelial cell response to treatment, the role of neuropilin-1 in wound healing or quantification of vessel formation in mice tumors. The methods include image enhancement, cell segmentation, detection and quantification of phenotypic changes. The image analysis pipelines were built in the open source software CellProfiler (Figure 12).
|
Alexandra Pacureanu, Carolina Wählby, Martin Simonsson
Partners: Karin Forsberg-Nilsson, Tanja Paavilainen, Soumi Kundu, Grzegorz Wicher, Lisa Rebello, Anqi Xiong, Dept. of Immunology, Genetics and Pathology, Rudbeck Laboratory, SciLife Lab Uppsala
Funding: SciLife Lab Uppsala
Period: 1210-
Abstract: Neural stem cells are the building blocks of the nervous system. In the view of finding better treatments for neurodegenerative diseases and for deeper understanding of mammalian development, our collaborators are investigating how neural stem cells proliferate and differentiate and which factors govern these processes. For theses studies, thousands of images of cell cultures need to be quantitatively analysed, in order to determine for example how effective are various techniques for control of the stem cells differentiation. Based on CellProfiler and CellProfiler Analyst, we have developed methods for automatic analysis of these images.
Ida-Maria Sintorn, Carolina Wählby
Partners: Mark Nicholas, Mats Josefson, AstraZeneca, Mölndal, Sweden
Funding: Pre-study grant from AIMDay Image, UU Innovation
Period: 1204-
Abstract: It is known qualitatively that microstructural differences in solid dosage forms (e.g. tablets and inhalation powders) affect the performance of the medication. The microstructural differences are differences in the spatial distribution of active and inactive compounds. The aim of this project is to characterize these microstructural differences in order to determine whether imaging techniques such as CLSM (confocal laser scanning microscopy), wide-field fluorescence microscopy, and TOF-SIMS (Time-Of-Flight Secondary Ion Mass Spectroscopy) can reveal quantifiable differences in structure. AstraZeneca researchers will then test if the quantified structural differences can be correlated with performance measures such as drug release. If successful, this research could lead to methods for drug quality control as well as an improved design of formulation and production protocols for AstraZeneca.
Ida-Maria Sintorn, Robin Strand
Partners: Ingela Parmryd, Dept. of Medical Cell Biology, UU; Jeremy Adler, Dept. Of Immunology, Genetics and Pathology, UU
Funding: TN-faculty, UU; S-faculty, SLU; VINNMER programme, Swedish Governmental Agency for Innovation Systems
Period: 1101-
Abstract: A cell surface is a highly irregular and rough. The surface
topography is however usually ignored in current models of the
plasma membrane, which are based on 2D observations of diffusion
that really occurs in 3D. In this project we model diffusion on
non-flat surfaces to explain biological processes occurring on the
cellsurface. During 2012, an abstract was accepted for the Biophysical Society 2013 Annual Meeting, Philadelphia, PA.
Andreas Kårsnäs, Robin Strand, Carolina Wählby, Ewert Bengtsson
Partners: Visiopharm, Hørsholm, Denmark; Clinical Pathology Division, Vejle hospital, Vejle, Denmark
Funding: NordForsk Private Public Partnership PhD Programme and Visiopharm
Period: 0909-
Abstract: The results of analyses of tissue biopsies by pathologists are crucial for breast cancer patients. In particular, the precision of a patient's prognosis, and the ability to predict the consequences of various treatment opportunities before actually exposing the cancer patient, depend on the detection and quantification of biomarkers in tissue sections by microscopy. Experience from the last decade has revealed that manual detection and quantification of biomarkers by microscopy of tissue biopsies is highly dependent on the competencies and stamina of the individual pathologist. The aim of the present PhD project is to develop software-based algorithms that can facilitate the workflow and ensure objective and more precise results of the quantitative microscopy procedures in breast cancer.
During 2012, we have, in collaboration with the Human Protein Atlas project, worked on a project for verifying antibodies by comparing staining patterns in immune-stained histological images. We made a comparison of different methods for classifying staining patterns in histology. This work was presented at MICCAI'12 in Nice. We have also presented the vectorial minimum barrier distance, a new method for computing gray-weighted distance transforms while incorporating vectorial data, at ICPR'12 in Tsukuba, Japan. Finally, we started a new project aimed at developing a new method for registering histological images of consecutive sections with different staining.
Bettina Selig, Cris Luengo
Partners: Bernd Rieger, Quantitative Imaging Group, Delft University
of Technology, Netherlands; Koen Vermeer, Eye Hospital Rotterdam,
Netherlands
Funding: S-faculty, SLU
Period: 1103-
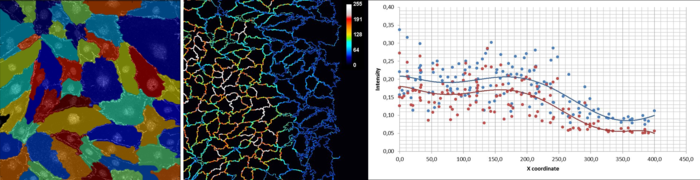
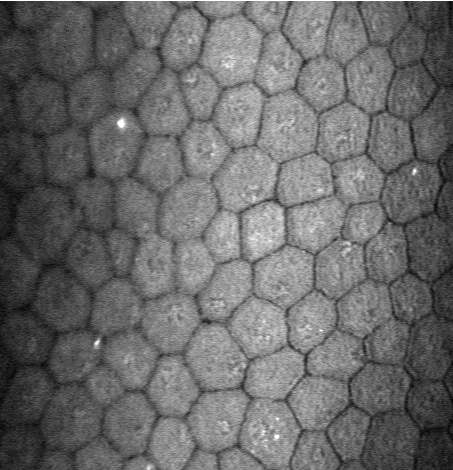
Abstract: In many corneal studies, endothelial cell density and morphology is
used to assess the quality of the cornea. Based on these parameters,
important therapeutic decisions are made. The endothelium may be
imaged by specular microscopy or by confocal scanners and measurements
can be obtained manually, automatically with manual corrections or
fully automatically with current software (e.g., Nidek's Navis).
Unfortunately, the results of the automatic mode are insufficient (see
Figure 13) when the image quality is affected or when irregular
shaped endothelial cells are present. In this project, we are
developing a new segmentation method, using stochastic watershed, that
enables a better estimation of endothelial quality.
 a |

b |
 c |
Ingrid Carlbom, Ewert Bengtsson, Jimmy Azar, Carolina Wählby
Partners: Christer Busch, Dept. of Genetics and Pathology, University Hospital; Marene Landström, Medical Biosciences, Umeå University.
Funding: Vetenskapsrådet (2009-5418): 2400000 SEK
Period: 1001-1212
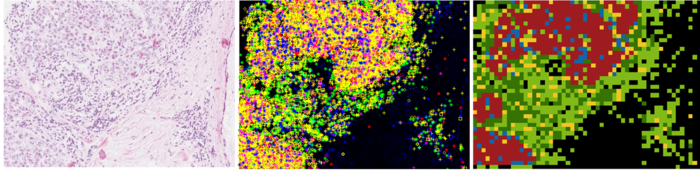
Abstract: Pathologists rely on multiple, contrasting stains to analyze tissue samples, but histological stains were developed for visual analysis and are not always ideal for automatic analysis. However, the choice of histological stain for automatic analysis should be based on its classification and clustering performance which are indicators of the performance of automatic segmentation of tissue into morphological components which in turn may be the basis for diagnosis. This year we explored a methodology for assessing the performance of both supervised and unsupervised classification on a given dataset. For supervised classification we measured the nonlinear support vector machines error rate, and for unsupervised classification we use the Rand index and the F-measure to assess the clustering results of a Gaussian mixture model based on expectation-maximization. Finally we investigated class separability measures based on scatter criteria.
Our data comprised adjacent sections of prostate tissue from radical prostatectomies prepared by the Department of Pathology at the Uppsala University Hospital. The sections were stained with 13 different histological stains; three of the stains, hematoxylin-eosin, Mallory's trichrome, and Sirius-hematoxylin, are shown in Figure 14. While the 13 stains are typically not all used for prostate cancer malignancy grading, they were included for illustration purposes.
We are interested in three stain / tissue combinations, or classes, namely nuclei, stroma, and cytoplasm. For the supervised case, we required a training set constituting the ground truth as defined by a pathologist, and this was obtained by the manual selection of regions in each of the stained images and by the labeling of each region according to its class. We trained and optimized a support vector classifier and obtained the classification error using ten-fold cross-validation. We chose the support vector classifier with a radial basis function, as it provides a significant range of complexity that may be controlled by optimizing the kernel and regularization parameters based on the cross-validated classification error.
For the unsupervised case, we assessed the clustering performance of the Gaussian mixture model by comparing the cluster labels with those of the ground truth, using precision and recall. We use the Gaussian mixture model, as it is not constrained by the assumption of spherical clusters as are the k-means and fuzzy c-means. For cluster separability we use two measures, the Fisher Criterion and the Mahalanobis distance.
We demonstrated that for a specific tissue type, in this case prostate tissue, the same five stains perform consistently better than the others according to objective error criteria for both the supervised and the unsupervised cases, as well as for the separability measures. The blind classification using the Gaussian mixture model and the Rand and F1-measures, show that Mallory Trichrome and Sirius-Htx+ outperform other stains in terms of clustering accuracy.
Based on these results we are creating a training data set for our continued work on prostate tissue segmentation. We selected the top performing stains in our ranking based on clustering efficacy, Sirius-Htx+ and Mallory Trichrome, and the widely used Hematoxylin-eosin as reference. We are creating new TMAs from the existing 180 patients in the Uppsala cohorts, where the tissue will be three consecutive sections from each patient, stained by the three stains, respectively. Since the tissue sections will be consecutive, they will be morphologically nearly equivalent, which will allow us to select an optimal stain based on the segmentation results.
Ida-Maria Sintorn
Partner: Vironova AB, Stockholm
Funding: VINNMER programme, Swedish Governmental Agency for Innovation Systems
Period: 0801-1207
Abstract: Electron Microscopy allows for studying the shape and morphology of biological particles such as viruses at the nm level. This means for example that structural differences between virus maturation stages, related virus species, wild type virus and virus treated with a potential drug or a small molecule can be analyzed. Both external (shape and protein patterns on the virus surface) and internal structural differences can be analyzed. In this project methods for efficiently identifying and quantifying such structural differences are developed. This Project is related to the Project (33) and a paper about a new texture measure based on a filtering approach of Zernike Moments was presented at ICPR in Tsukuba, in November.
Gustaf Kylberg, Ida-Maria Sintorn, Ewert Bengtsson, Gunilla Borgefors
Partner: Vironova AB; Delong Instruments, Brno, Czech Republic; Ali Mirazimi, Kjell-Olof Höglund, Centre for Microbiological Preparedness; Swedish Institute for Infectious Disease Control (SMI)
Funding: 2008-2011 Swedish Civil Contingencies Agency (MSB); Swedish Defense Materiel Administration (FMV); Swedish Agency for Innovative Systems (VINNOVA). 2011- Eurostar project E!6143
Period: 0801-
Abstract: Transmission electron microscopy (TEM) is an important virus diagnostic tool. The main drawback is that an expert in virus appearance in electron microscopy needs to perform the analysis at the microscope, an often very time consuming task.
The project aim is to develop methods for a multi-scale analysis at the microscope to automatically acquire high magnification focused images of possible virus particles. This is an important step towards automating the virus identification process and thereby creating a rapid, objective, and user independent virus diagnostic system. By introducing the multi-scale approach the search area where high magnification images need to be acquired is estimated to be reduced with more than 99.99%.
Delong Instruments has joined the project. They will develop a novel bench-top low-voltage TEM where the methods for automated acquisition will be incorporated. This work will intensify the development of methods for the automatic acquisition of images including methods for automatic focusing and astigmatism correction.
Partner: Vironova AB; Delong Instruments, Brno, Czech Republic; Ali Mirazimi, Kjell-Olof Höglund, Centre for Microbiological Preparedness; Swedish Institute for Infectious Disease Control (SMI)
Funding: 2008-2011 Swedish Civil Contingencies Agency (MSB); Swedish Defense Materiel Administration (FMV); Swedish Agency for Innovative Systems (VINNOVA). 2011- Eurostar project E!6143
Period: 0801-
Abstract: This project aims at automating the virus identification process in high resolution TEM images. This, in combination with Project 32 create a rapid, objective, and user independent virus diagnostic system. The identification task consists of method development for segmenting virus particles with different shapes and sizes and extracting descriptive features of both shape and texture to enable the classification into virus species. A paper describing such a segmentation method was published in Journal of Microscopy in early 2012. Texture features such as variants of Local Binary Patterns are being evaluated on virus textures as well as other texture datasets to get a deeper understanding of the discriminant power of the features under different conditions.
Work on texture features based on Zernike moments was presented at the 21st International Conference on Pattern Recognition (ICPR 2012) in Tsukuba, Japan. Figure 15 shows Zernike moments from order 1 to 6.
Azadeh Fakhrzadeh, Cris Luengo, Gunilla Borgefors
Partners: Ellinor Spörndly-Nees, Lena Holm, Dept. of Anatomy, Physiology and Biochemistry, SLU
Funding: SLU (KoN)
Period: 1009-
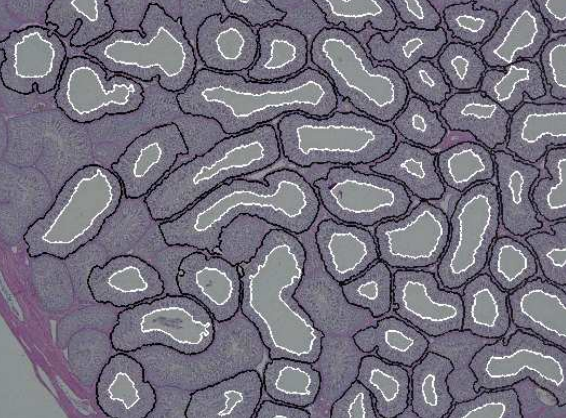
Abstract: Reproductive toxicology is the study of chemicals and their effects on the reproductive system of humans and animals. In reproductive toxicology, there is a strong need to detect structural differences in organs that often have both a complex microscopic structure and function. This problem is
further complicated because standard techniques are based on the examination of two-dimensional sections of a three-dimensional structure. The aim of this project is to develop methods to objectively describe microscopic structures of male reproductive organs and to test these in reproductive toxicology research. The project is comparative and includes studies of organs from rooster and mink. We are developing automatic and interactive methods to analyze the relevant structures in the histology images of testis. We have constructed an automatic method to delineate the seminiferous tubule border and lumen. We use a level set based active contour method to delineate the
lumen border. Detecting the border of tubules is a challenging problem. The tubules have low contrast borders and they appears broken in some parts. We use a method based on geodistic distance transform to separate the clustered tubules. We compute the evolving curve inside a connected component using the geodesic distance transform. The lumen will serve as initialization for the
evolving curve. The length of the curve increases until it reaches the border of object, then it starts to decrease, and as soon as the curve enters another tubule, the length of curve increases again. We will thus have a clear minimum where the tubules connect to each other. The result of segmentation is shown in Figure 16. This is published in DICTA 2012.
|
Hamid Sarve, Gunilla Borgefors
Partners: Carina Johansson, The Sahlgrenska Academy, Göteborg
Funding: S-faculty, SLU
Period: 0503-
Abstract: With an aging and increasingly osteoporotic population, bone implants are becoming more important to ensure the quality of life. In order to evaluate how tissue and bone reacts on implants, the interface at the implant and tissue must be studied. The aim of this project is to develop automatic image analysis methods for evaluating images of the interface region of tissue and implant. The images come both from traditional light microscopy that gives detailed 2D images and from synchrotron micro-CT (SRµCT) images that gives less detailed but fully 3D images. The project has developed image analysis methods for both modalities separately and for comparing them. We have also developed new techniques for interactive 3D visualization of bone anchored implants in order to facilitate the understanding of the mechanisms of implant integration. The new methods were applied on real clinical data; a case study involving retrieved human oral implants. All methods were collected in the thesis of Sarve, that was presented in 2011. This year, a semi-popular summary was published in Tandläkartidningen, the journal of the Swedish dentists.
|